File MIES_AnalysisFunctions_PatchSeq.ipf¶
PSQ Analysis functions for patch sequence
The stimsets with the analysis functions attached are executed in the following order:
PSQ_PipetteInBath
PBPSQ_SealEvaluation
SEPSQ_AccessResistanceSmoke
ARPSQ_TrueRestingMembranePotential
VMPSQ_DaScale (sub threshold)
DA (sub)PSQ_SquarePulse (long pulse)
SPPSQ_Rheobase (long pulse)
RBPSQ_DaScale (supra threshold)
DA (supra)PSQ_DaScale (adaptive threshold)
DA (adaptive)PSQ_SquarePulse (short pulse)
SPPSQ_Rheobase (short pulse)
RBPSQ_Ramp
RAPSQ_Chirp
CR
See GetAbbreviationForAnalysisFunction() for a generating the abbreviation of each analysis function.
The Patch Seq analysis functions store various results in the labnotebook.
For orientation the following table shows their relation. All labnotebook keys can and should be created via CreateAnaFuncLBNKey().
Naming constant |
Description |
Unit |
Labnotebook |
Analysis function |
Per Chunk? |
Headstage dependent? |
|---|---|---|---|---|---|---|
PSQ_FMT_LBN_SPIKE_DETECT |
The required number of spikes were detected on the sweep |
On/Off |
Numerical |
SP, RB, RA, DA (Supra) |
No |
Yes |
PSQ_FMT_LBN_SPIKE_POSITIONS |
Spike positions |
ms |
Numerical |
RA, VM |
No |
Yes |
PSQ_FMT_LBN_SPIKE_COUNT |
Spike count |
(none) |
Numerical |
DA (Supra) |
No |
Yes |
PSQ_FMT_LBN_STEPSIZE |
Current DAScale step size |
(none) |
Numerical |
SP, RB |
No |
No |
PSQ_FMT_LBN_STEPSIZE_FUTURE |
Future DAScale step size |
(none) |
Numerical |
RB |
No |
No |
PSQ_FMT_LBN_RB_DASCALE_EXC |
Range for valid DAScale values is exceeded |
On/Off |
Numerical |
RB |
No |
Yes |
PSQ_FMT_LBN_DASCALE_OOR |
Future DAScale value is out of range |
On/Off |
Numerical |
CR,XXX |
Yes |
Yes |
PSQ_FMT_LBN_RB_LIMITED_RES |
Failed due to limited DAScale resolution |
On/Off |
Numerical |
RB |
No |
Yes |
PSQ_FMT_LBN_FINAL_SCALE |
Final DAScale of the given headstage, only set on success |
(none) |
Numerical |
SP, RB |
No |
No |
PSQ_FMT_LBN_SPIKE_DASCALE_ZERO |
Sweep spiked with DAScale of 0 |
On/Off |
Numerical |
SP |
No |
No |
PSQ_FMT_LBN_INITIAL_SCALE |
Initial DAScale |
(none) |
Numerical |
RB, CR |
No |
No |
PSQ_FMT_LBN_RMS_SHORT |
Short RMS baseline value |
V |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_RMS_SHORT_PASS |
Short RMS baseline QC result |
On/Off |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_RMS_SHORT_THRESHOLD |
Short RMS baseline threshold |
V |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
No |
Yes |
PSQ_FMT_LBN_RMS_LONG |
Long RMS baseline value |
V |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_RMS_LONG_PASS |
Long RMS baseline QC result |
On/Off |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_RMS_LONG_THRESHOLD |
Long RMS baseline threshold |
V |
Numerical |
DA, RB, RA, CR, SE, VM, AR |
No |
Yes |
PSQ_FMT_LBN_TARGETV |
Target voltage baseline |
Volt |
Numerical |
DA, RB, RA, CR |
Yes |
Yes |
PSQ_FMT_LBN_TARGETV_PASS |
Target voltage baseline QC result |
On/Off |
Numerical |
DA, RB, RA, CR |
Yes |
Yes |
PSQ_FMT_LBN_TARGETV_THRESHOLD |
Target voltage threshold |
V |
Numerical |
DA, RB, RA, CR |
No |
Yes |
PSQ_FMT_LBN_LEAKCUR |
Leak current |
Amperes |
Numerical |
PB, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_LEAKCUR_PASS |
Leak current QC result |
On/Off |
Numerical |
PB, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_AVERAGEV |
Average voltage |
Volt |
Numerical |
VM |
Yes |
Yes |
PSQ_FMT_LBN_CHUNK_PASS |
Which chunk passed/failed baseline QC |
On/Off |
Numerical |
DA, RB, RA, CR, PB, SE, VM, AR |
Yes |
Yes |
PSQ_FMT_LBN_BL_QC_PASS |
Pass/fail state of the complete baseline |
On/Off |
Numerical |
DA, RB, RA, CR, PB, SE, VM, AR |
No |
Yes |
PSQ_FMT_LBN_SWEEP_PASS |
Pass/fail state of the complete sweep |
On/Off |
Numerical |
DA, SP, RA, CR, PB, SE, VM, AR |
No |
No |
PSQ_FMT_LBN_SWEEP_EXCEPT_BL_PP_PASS |
Pass/fail state of the complete sweep wo post pulse baseline |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_SET_PASS |
Pass/fail state of the complete set |
On/Off |
Numerical |
DA, RB, RA, SP, CR, PB, SE, VM, AR |
No |
No |
PSQ_FMT_LBN_SAMPLING_PASS |
Pass/fail state of the sampling interval check |
On/Off |
Numerical |
DA, RB, RA, SP, CR, PB, SE, VM, AR |
No |
No |
PSQ_FMT_LBN_PULSE_DUR |
Pulse duration as determined experimentally |
ms |
Numerical |
RB, DA, CR |
No |
Yes |
PSQ_FMT_LBN_SPIKE_PASS |
Pass/fail state of the spike search (No spikes → Pass) |
On/Off |
Numerical |
CR, VM |
No |
Yes |
PSQ_FMT_LBN_DA_fI_SLOPE |
Fitted slope in the f-I plot |
% Hz/pA |
Numerical |
DA (Supra) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FI_NEG_SLOPE_PASS |
Fitted slop in the f-I plot is negative |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_fI_SLOPE_REACHED_PASS |
Fitted slope in the f-I plot exceeds target value |
On/Off |
Numerical |
DA (Supra, Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_FI_OFFSET |
Fitted offset in the f-I plot |
Hz |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FI_SLOPE |
Fitted slope in the f-I plot |
% Hz/pA |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FI_OFFSET_DASCALE |
Fitted offset in the f-I plot for DAScale estimation |
Hz |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FI_SLOPE_DASCALE |
Fitted slope in the f-I plot for DAScale estimation |
% Hz/pA |
Numerical |
DA (Supra) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FREQ |
AP frequency |
Hz |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FREQ |
AP frequencies from previous sweep reevaluation |
Hz |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_DASCALE |
DAScale values from previous sweep reevaluation |
(none) |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FI_OFFSETS |
Offsets in the f-I plot from previous sweep reevaluation |
Hz |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FI_SLOPES |
Slopes in the f-I plot from previous sweep reevaluation |
% Hz/pA |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FI_NEG_SLOPES_PASS |
Fitted slop in the f-I plot is negative |
On/Off |
Textual |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_RSA_FI_OFFSETS_DASCALE |
Offsets in the f-I plot from previous sweep reevaluation |
Hz |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FI_SLOPES_DASCALE |
Slopes in the f-I plot from previous sweep reevaluation |
% Hz/pA |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FI_SLOPES_PASS |
Slope QCs in the f-I plot from previous sweep reevaluation |
On/Off |
Textual |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_RSA_VALID_SLOPE_PASS |
Valid slope from f-I plot QC of supra sweep reevaluation |
On/Off |
Numerical |
None |
No |
Yes |
PSQ_FMT_LBN_DA_AT_RSA_FILLIN_PASS |
Acquired due to fillin from previous sweep reevaluation |
On/Off |
Numerical |
None |
No |
No |
PSQ_FMT_LBN_DA_AT_MAX_SLOPE |
Maximum encountered f-I plot slope in the SCI |
% Hz/pA |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_VALID_SLOPE_PASS |
Valid slope from f-I plot QC |
On/Off |
Numerical |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_NEG_SLOPE_FILLIN |
Fillin for negative f-I slope passing is ongoing |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_ENOUGH_FI_POINTS_PASS |
Enough f-I pairs for line fit QC |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_FUTURE_DASCALES |
DAScale values left to acquire |
(none) |
Textual |
DA (Adapt) |
No |
Yes |
PSQ_FMT_LBN_DA_AT_FUTURE_DASCALES_PASS |
Measured all DAScale values |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_MIN_DASCALE_NORM |
Minimum DAScale step width (normalized) |
(none) |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_MAX_DASCALE_NORM |
Maximum DAScale step width (normalized) |
(none) |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_AT_FILLIN_PASS |
Acquired due to fillin |
On/Off |
Numerical |
DA (Adapt) |
No |
No |
PSQ_FMT_LBN_DA_OPMODE |
Operation Mode: One of PSQ_DS_SUB/PSQ_DS_SUPRA/PSQ_DS_ADAPT |
(none) |
Textual |
DA |
No |
No |
PSQ_FMT_LBN_CR_INSIDE_BOUNDS |
AD response is inside the given bands |
On/Off |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_RESISTANCE |
Calculated resistance from DAScale sub threshold |
Ohm |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_BOUNDS_ACTION |
Action according to min/max positions |
(none) |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_BOUNDS_STATE |
Upper and Lower bounds state according to min/max pos. |
(none) |
Textual |
CR |
No |
No |
PSQ_FMT_LBN_CR_SPIKE_CHECK |
Spike check was enabled/disabled |
(none) |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_INIT_UOD |
Initial user onset delay |
ms |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_INIT_LPF |
Initial MCC low pass filter |
Hz |
Numerical |
CR |
No |
No |
PSQ_FMT_LBN_CR_STIMSET_QC |
Stimset valid/invalid QC state |
On/Off |
Numerical |
CR |
No |
No |
FMT_LBN_ANA_FUNC_VERSION |
Integer version of the analysis function |
(none) |
Numerical |
All |
No |
Yes |
PSQ_FMT_LBN_PB_RESISTANCE |
Pipette Resistance |
Ohm |
Numerical |
PB |
No |
No |
PSQ_FMT_LBN_PB_RESISTANCE_PASS |
Pipette Resistance QC |
On/Off |
Numerical |
PB |
No |
No |
PSQ_FMT_LBN_SE_TP_GROUP_SEL |
Selected Testpulse groups: One of Both/First/Second |
(none) |
Textual |
SE |
No |
No |
PSQ_FMT_LBN_SE_RESISTANCE_A |
Seal Resistance of TPs in group A |
Ω |
Numerical |
SE |
No |
No |
PSQ_FMT_LBN_SE_RESISTANCE_B |
Seal Resistance of TPs in group B |
Ω |
Numerical |
SE |
No |
No |
PSQ_FMT_LBN_SE_RESISTANCE_MAX |
Maximum Seal Resistance of TPs in both groups |
Ω |
Numerical |
SE |
No |
No |
PSQ_FMT_LBN_SE_RESISTANCE_PASS |
Seal Resistance QC |
On/Off |
Numerical |
SE |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG |
Average voltage of all baseline chunks |
Volt |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG_ADIFF |
Average voltage absolute difference |
Volt |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG_ADIFF_PASS |
Average voltage absolute difference QC |
On/Off |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG_RDIFF |
Average voltage relative difference |
(none) |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG_RDIFF_PASS |
Average voltage relative difference QC |
On/Off |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_VM_FULL_AVG_PASS |
Full average voltage QC result |
On/Off |
Numerical |
VM |
No |
No |
PSQ_FMT_LBN_AR_ACCESS_RESISTANCE |
Access resistance of all TPs in the stimset |
Ω |
Numerical |
AR |
No |
No |
PSQ_FMT_LBN_AR_ACCESS_RESISTANCE_PASS |
Access resistance QC |
On/Off |
Numerical |
AR |
No |
No |
PSQ_FMT_LBN_AR_STEADY_STATE_RESISTANCE |
Steady state resistance of all TPs in the stimset |
Ω |
Numerical |
AR |
No |
No |
PSQ_FMT_LBN_AR_RESISTANCE_RATIO |
Ratio of access resistance to steady state |
(none) |
Numerical |
AR |
No |
No |
PSQ_FMT_LBN_AR_RESISTANCE_RATIO_PASS |
Ratio of access resistance to steady state QC |
On/Off |
Numerical |
AR |
No |
No |
PSQ_FMT_LBN_ASYNC_PASS |
Combined alarm state of all async channels |
On/Off |
Numerical |
All |
No |
No |
LBN_DELTA_I |
Delta current in pulse |
Amperes |
Numerical |
RV, AD, DA (Sub) |
No |
Yes |
LBN_DELTA_V |
Delta voltage in pulse |
Volts |
Numerical |
RV, AD, DA (Sub) |
No |
Yes |
LBN_RESISTANCE_FIT |
Fitted resistance from pulse |
Ohm |
Numerical |
RV, AD, DA (Sub) |
No |
Yes |
LBN_RESISTANCE_FIT_ERR |
Error of fitted resistance from pulse |
Ohm |
Numerical |
RV, AD, DA (Sub) |
No |
Yes |
LBN_AUTOBIAS_TARGET_DIAG |
Autobias target voltage from dialog |
mV |
Numerical |
RV |
No |
Yes |
Query the standard STIMSET_SCALE_FACTOR_KEY entry from labnotebook for getting the DAScale.
The following table lists the user epochs which are added during data acquisition:
Tags |
Short Name |
Analysis function |
Description |
Level |
|---|---|---|---|---|
Name=Baseline Chunk;Index=x |
U_BLCx |
DA, RB, RA, CR, SE, VM, AR |
Baseline QC evaluation chunks |
-1 |
Name=Baseline Chunk QC Selection;Index=x |
U_BLSx |
SE, VM, AR |
Selects the chunk for Baseline QC evaluation |
-1 |
Name=DA Suppression |
U_RA_DS |
RA |
DA was suppressed in this time interval |
-1 |
Name=Unacquired DA data |
U_RA_UD |
RA |
Interval of unacquired data |
-1 |
Type=Testpulse Like;Index=x |
U_TPx |
PB, SE, AR |
Testpulse like region in stimset |
-1 |
Type=Testpulse Like;SubType=Baseline;Index=x |
U_TPx_B0 |
PB, SE, AR |
Pre pulse baseline of testpulse |
-1 |
Type=Testpulse Like;SubType=Pulse;Amplitude=y;Index=x |
U_TPx_P |
PB, SE, AR |
Pulse of testpulse |
-1 |
Type=Testpulse Like;SubType=Baseline;Index=x |
U_TPx_B1 |
PB, SE, AR |
Post pulse baseline of testpulse |
-1 |
Type=Chirp Cycle Evaluation |
U_CR_CE |
CR |
Evaluation chunk for bounds state |
-1 |
Type=Chirp Spike Evaluation |
U_CR_SE |
CR |
Evaluation chunk for spike check |
-1 |
The tag entry Index=x is a zero-based index, which tracks how often the specific type of user epoch appears. So for different
epoch types duplicated index entries are to be expected.
See also Time specialities.
Textual results entries stored by the sweep formula store operation with the names Sweep Formula store [XXX]:
Name |
Analysis function |
|---|---|
Steady state resistance |
PB |
Steady state resistance (group A) |
PB |
Steady state resistance (group B) |
PB |
Average U_BLS0 |
VM |
Average U_BLS1 |
VM |
Steady state resistance |
AR |
Peak resistance |
AR |
Type constants for PSQ_DS_GetLabnotebookData
-
static const double PSQ_DS_FI_SLOPE = 0x1¶
-
static const double PSQ_DS_FI_SLOPE_DASCALE = 0x2¶
-
static const double PSQ_DS_FI_SLOPE_REACHED_PASS = 0x3¶
-
static const double PSQ_DS_FI_NEG_SLOPE_PASS = 0x4¶
-
static const double PSQ_DS_FI_OFFSET = 0x5¶
-
static const double PSQ_DS_FI_OFFSET_DASCALE = 0x6¶
-
static const double PSQ_DS_SWEEP_PASS = 0x7¶
-
static const double PSQ_DS_SWEEP = 0x8¶
-
static const double PSQ_DS_APFREQ = 0x9¶
-
static const double PSQ_DS_DASCALE = 0xa¶
-
static const double PSQ_DS_SWEEP_EXCEPT_BL_PP_PASS = 0xb¶
-
static const double PSQ_DS_FILLIN_PASS = 0xc¶
Functions
-
variable PSQ_GetPulseSettingsForType(variable type, PSQ_PulseSettings *s)¶
Fills
saccording to the analysis function type.
-
static wave PSQ_GetPulseDurations(string device, variable type, variable sweepNo, variable totalOnsetDelay, variable forceRecalculation = defaultValue)¶
Return the pulse durations from the labnotebook or calculate them before if required in ms. For convenience unused headstages will have 0 instead of NaN in the returned wave.
We recalculate the durations on every sweep again as we can not assume that it is constant for the whole stimulus set.
-
static wave PSQ_DeterminePulseDuration(string device, variable sweepNo, variable type, variable totalOnsetDelay)¶
Determine the pulse duration on each headstage.
Returns the labnotebook wave as well.
-
wave PSQ_DeterminePulseDurationFinder(wave statusHS, WaveRefWave allSingleDA, variable type, variable totalOnsetDelay)¶
-
static std::tuple<variable, variable, variable> PSQ_GetBaselineThresholds(string params)¶
Return the baseline RMS short/long and target V thresholds in mV.
-
static variable PSQ_StoreThresholdsInLabnotebook(string device, variable type, variable sweepNo, variable headstage, variable rmsShortThreshold, variable rmsLongThreshold, variable targetVThreshold)¶
-
static variable PSQ_EvaluateBaselinePassed(string device, variable type, variable sweepNo, variable headstage, variable chunk, variable ret)¶
-
static std::tuple<variable, variable> PSQ_EvaluateBaselineChunks(string device, variable type, AnalysisFunction_V3 *s)¶
-
std::tuple<variable, variable> PSQ_GetBaselineChunkTimes(variable chunk, PSQ_PulseSettings *ps, variable totalOnsetDelayMS, WaveOrNull durations)¶
Calculates the baseline chunk times.
-
static variable PSQ_EvaluateBaselineProperties(string device, AnalysisFunction_V3 *s, variable type, variable chunk, variable fifoInStimsetTime, variable totalOnsetDelay)¶
Evaluate one chunk of the baseline.
- Parameters:
device – device
s – AnalysisFunction_V3 struct
type – analysis function type, one of PatchSeqAnalysisFunctionTypes
chunk – chunk number,
chunk == 0-> Pre pulse baseline chunk,chunk >= 1-> Post pulse baseline for traditional baseline evaluation if PSQ_PulseSettings::usesBaselineChunkEpochs is false.fifoInStimsetTime – Fifo position in ms relative to the start of the stimset (therefore ignoring the totalOnsetDelay)
totalOnsetDelay – total onset delay in ms
- Returns:
pre pulse baseline: 0 if the chunk passes, one of the possible AnalysisFuncReturnTypesConstants values otherwise post pulse baseline: 0 if the chunk passes, PSQ_BL_FAILED if it does not pass generic baseline: 0 if the chunk passes, PSQ_BL_FAILED if it does not pass
-
variable PSQ_AddBaselineEpoch(wave epochWave, variable DAC, variable chunk, variable chunkStartTimeMax, variable chunkLengthTime)¶
Add base line epoch to user epochs.
- Parameters:
epochWave – 4D epochs wave
DAC – DAC channel number
chunk – number of evaluated chunk
chunkStartTimeMax – start time of chunk in ms
chunkLengthTime – time length of chunk in ms
-
static variable PSQ_GetNumberOfChunks(string device, variable sweepNo, variable headstage, variable type, variable sampleIntervalAD)¶
Return the number of chunks.
A chunk is PSQ_BL_EVAL_RANGE [ms] of baseline.
For calculating the number of chunks we ignore the one chunk after the pulse which we don’t evaluate!
-
static variable PSQ_Calculate(wave wv, variable column, variable startTime, variable rangeTime, variable method)¶
-
static variable PSQ_NumAcquiredSweepsInSet(string device, variable sweepNo, variable headstage)¶
Return the number of already acquired sweeps from the given stimset cycle.
-
variable PSQ_NumPassesInSet(wave numericalValues, variable type, variable sweepNo, variable headstage)¶
Return the number of passed sweeps in all sweeps from the given stimset cycle.
-
static variable PSQ_GetDAStimsetLength(string device, variable headstage)¶
Return the DA stimset length in ms of the given headstage.
- Returns:
stimset length or -1 on error
-
wave PSQ_CreateOverrideResults(string device, variable headstage, variable type, string opMode = defaultValue)¶
Create the overrides results wave for CI testing.
Rows:
Only one
Cols:
sweeps/steps
Layers:
0: x position in ms where the spike is in each sweep/step For convenience the values
0always means no spike and1spike detected (at the appropriate position).1: async channel QC
Rows:
chunk indizes
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: x position in ms where the spike is in each sweep/step For convenience the values
0always means no spike and1spike detected (at the appropriate position).2: async channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
PSQ_DA_SCALE (Sub and Supra):
Rows:
chunk indizes
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: x position in ms where the spike is in each sweep/step For convenience the values
0always means no spike and1spike detected (at the appropriate position).2: Number of spikes
3: Async channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
PSQ_DA_SCALE (Adaptive):
Rows:
chunk indizes
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: APFrequency
2: Async channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
JSON-Wavenote:
”APFrequenciesRhSuAd”: List of frequencies from the previous passing rheobase/supra/adaptive SCI
”DAScalesRhSuAd”: List of DAScale values from previous passing rheobase/supra/adaptive SCI
”PassingSupraSweep”: Sweep with passing supra set QC
”PassingRheobaseSweep”: Sweep with passing rheobase set QC
”PassingRhSuAdSweeps”: List of passing rheobase/supra/adaptive SCI sweeps
”PassingAdaptiveSweepFromFailingSet”: Passing adaptive sweeps from failing sets from previous SCIs (written by PSQ_DaScale)
Rows:
chunk indizes
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: maximum of AD data in chirp region [first row only, others are ignored], use NaN to use the real values
2: minimum of AD data in chirp region [first row only, others are ignored], use NaN to use the real values
3: passing spike check in chirp region or not [first row only, others are ignored]
4: async channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
Rows:
chunk indizes
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: averaged steady state resistance [MΩ]
2: async channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
Rows:
chunk indizes (only selected chunks via user epoch, sorted by ascending chunk start time)
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: Resistance A [MΩ]
2: Resistance B [MΩ]
3: Async Channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
Rows:
chunk indizes (only selected chunks via user epoch, sorted by ascending chunk start time)
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: number of spikes (QC passes with 0)
2: average voltage [mV]
3: async channel QC
Chunks (only for row/layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC (unused)
3: leak current baseline QC (unused)
Rows:
chunk indizes (only selected chunks via user epoch, sorted by ascending chunk start time)
Cols:
sweeps/steps
Layers:
0: 1 if the chunk has passing baseline QC or not
1: Access Resistance [MΩ]
2: Steady State Resistance [MΩ]
3: Async Channel QC
Chunks (only for layer 0):
0: RMS short baseline QC
1: RMS long baseline QC
2: target voltage baseline QC
3: leak current baseline QC
-
static variable PSQ_StoreStepSizeInLBN(string device, variable type, variable sweepNo, variable stepsize, variable future = defaultValue)¶
Store the step size in the labnotebook.
-
static wave PSQ_SearchForSpikes(string device, variable type, wave sweepWave, variable headstage, variable offset, variable level, variable searchEnd = defaultValue, variable numberOfSpikesReq = defaultValue, wave spikePositions = defaultValue, variable *numberOfSpikesFound = defaultValue)¶
Search the AD channel of the given headstage for spikes from the pulse onset until the end of the sweep or
searchEndif given.- Parameters:
device – [in] device
type – [in] One of PatchSeqAnalysisFunctionTypes
sweepWave – [in] sweep wave with acquired data
headstage – [in] headstage in the range [0, NUM_HEADSTAGES[
offset – [in] offset in ms where the spike search should start, commonly the totalOnsetDelay
level – [in] set the level for the spike search
searchEnd – [in] [optional, defaults to inf] total length in ms of the spike search, relative to sweepWave start
numberOfSpikesReq – [in] [optional, defaults to one] number of spikes to look for Positive finite value: Return value is 1 iff at least
numberOfSpikeswere found Inf: Return value is 1 if at least 1 spike was foundspikePositions – [out] [optional] returns the position of the first
numberOfSpikesfound on success in msnumberOfSpikesFound – [out] [optional] returns the number of spikes found
- Returns:
labnotebook value wave suitable for ED_AddEntryToLabnotebook()
-
static variable PSQ_GetLastPassingLongRHSweep(string device, variable headstage, variable duration)¶
Return a sweep number of an existing sweep matching the following conditions.
Acquired with Rheobase analysis function
Set QC passed for this SCI
Pulse duration was longer than
durationSpiked
And as usual we want the last matching sweep.
- Returns:
existing sweep number or INVALID_SWEEP_NUMBER in case no such sweep could be found
-
static variable PSQ_IsSuitableFailingAdaptiveSweep(string device, variable sweepNo, variable headstage, string params)¶
Return the truth that sweepNo/headstage is from an adaptive SCI with special properties.
Special properties:
DAScale supra adaptive analysis function was run
failing set QC
all analysis parameters are the same
used same targetV autobias value
-
static wave PSQ_GetPreviousSetQCFailingAdaptive(string device, variable headstage, string params)¶
Return passing sweeps from failing adaptive sets.
Look into as many previous SCIs as given in the FailingAdaptiveSCIRange analysis parameter to return the passing sweeps where the SCI fullfills the properties listed at PSQ_IsSuitableFailingAdaptiveSweep()
-
variable PSQ_DS_IsValidMode(string str)¶
Return the truth that str is a valid PSQ_DAScale operation mode.
-
static variable PSQ_GetLastPassingDAScale(string device, variable headstage, string opMode)¶
Return the sweep number of the last sweep using the PSQ_DaScale() analysis function, where the set passes and was in the given mode.
-
static variable PSQ_GetLastPassingTrueRMP(string device, variable headstage)¶
Return the sweep number of the last sweep using the PSQ_TrueRestingMembranePotential() analysis function.
-
static variable PSQ_DS_GetDAScaleOffset(string device, variable headstage, string opMode)¶
Return the DAScale offset for PSQ_DaScale()
- Returns:
DAScale value in pA or NaN on error
-
static variable PSQ_FoundAtLeastOneSpike(string device, variable sweepNo)¶
Check if the given sweep has at least one “spike detected” entry in the labnotebook.
- Returns:
1 if found at least one, zero if none and
NaNif no such entry could be found
-
static variable PSQ_GetDefaultSamplingFrequency(string device, variable type)¶
-
variable PSQ_GetDefaultSamplingFrequencyForSingleHeadstage(string device)¶
Returns the sampling frequency in kHz of our supported NI and ITC hardware for one active headstage and no TTL channels.
-
static variable PSQ_CheckADSamplingFrequencyAndStoreInLabnotebook(string device, variable type, AnalysisFunction_V3 *s)¶
Return the QC state of the sampling interval/frequency check and store it also in the labnotebook.
-
static variable PSQ_DS_GatherEpochDuration(string setName, variable epochIndex)¶
Return the duration of the given epoch in the stimulus set.
Does not require epoch info, and also supports combine epochs which don’t have a “Duration” parameter.
-
static std::tuple<variable, variable> PSQ_DS_GetFrequencyEvalRangeForRhSuAd(string device, variable headstage, variable sweepNo)¶
-
static std::tuple<variable, variable> PSQ_DS_GetFrequencyEvalRange(string device, variable headstage, variable sweepNo)¶
-
static std::tuple<WAVE, WAVE> PSQ_DS_GatherFrequencyCurrentData(string device, variable sweepNo, variable headstage, wave sweeps, variable fromRhSuAd = defaultValue)¶
Use SweepFormula to gather the AP frequency and DAScale data from the given sweeps.
- Parameters:
device – DAC hardware device
sweepNo – current sweep
headstage – headstage
sweeps – wave of sweep numbers to gather the data from
fromRhSuAd – [optional, defaults to off] called for rheobase/supra/previous adaptive data (on) or adaptive data (off)
- Return values:
apfreq – AP frequency data
DAScale – DAScale data
-
static std::tuple<WaveDouble, WaveDouble, string> PSQ_DS_FitFrequencyCurrentData(string device, variable sweepNo, WaveOrNull apfreq, WaveOrNull DAScales, variable singleFit = defaultValue, variable writeEnoughPointsLBN = defaultValue)¶
Fit the AP frequency and DAScale data with a line fit and return the fit results (offsets and slopes)
- Parameters:
device – DAC hardware device
sweepNo – current sweep
apfreq – AP frequency data
DAScales – DAScale data
singleFit – [optional, defaults to false] if only a single fit should be done
writeEnoughPointsLBN – [optional, defaults to false] if the labnotebook entry for enough points should be written or not
- Return values:
fitOffset – offsets of all line fits
fitSlope – slopes of all line fits
errMsg – error message if both
fitOffsetandfitSlopeare null
-
static variable PSQ_DS_CreateSurveyPlotForUser(string device, variable sweepNo, variable headstage, variable fromRhSuAd = defaultValue)¶
-
std::tuple<variable, variable> PSQ_DS_GetDAScaleStepsNorm(string device, variable sweepNo, variable headstage)¶
-
static string PSQ_DS_AD_BuildFutureDAScaleEntry(string type, variable DAScale)¶
Build an entry like
type:DAScalefromtypeandDAScale
-
static std::tuple<string, variable> PSQ_DS_AD_ParseFutureDAScaleEntry(string DAScaleWithType)¶
Split an entry like
type:DAScaleintotypeandDAScale
-
static wave PSQ_DS_GatherDAScaleFillin(PSQ_DS_DAScaleParams *cdp, string type, wave apfreqParam, wave DAScalesParam, WaveTextOrNull futureDAScalesHistoric)¶
Calculate a list of DAScale values which need to be acquired in addition so that the AP frequency differences between consecutive points is smaller than
maxFrequencyChangePercent.- Returns:
combined dascale with one of the types from FutureDAScaleReason including the historic future DAScales at the front
-
static wave PSQ_DS_CalculateDAScale(PSQ_DS_DAScaleParams *cdp, wave numericalValues, wave textualValues, variable sweepNo, variable headstage, variable fromRhSuAd = defaultValue, variable alreadyDone = defaultValue)¶
-
static wave PSQ_DS_CalculateDAScaleFallback(PSQ_DS_DAScaleParams *cdp, variable sweepNo, string type, variable x, variable y)¶
-
static wave PSQ_DS_CalculateDAScaleForNegSlope(PSQ_DS_DAScaleParams *cdp, string type, variable x, variable y)¶
Calculate the new DAScale value after a negative fI slope.
- Parameters:
cdp – calculation parameters
type – one of FutureDAScaleReason
x – DAScale of last acquired sweep [pA]
y – frequency value of last acquired sweep [Hz]
-
static wave PSQ_DS_CalculateDAScalePlain(PSQ_DS_DAScaleParams *cdp, string type, variable a, variable m, variable x, variable y)¶
Calculate the new DAScale value for adaptive threshold using linear extrapolation.
- Parameters:
cdp – calculation parameters
type – one of FutureDAScaleReason
a – offset of the f-I straight line fit [Hz]
m – its slope [% Hz/pA]
x – DAScale of last acquired sweep [pA]
y – frequency value of last acquired sweep [Hz]
- Returns:
text wave with new DAScale value [pA] combined with one of the types from FutureDAScaleReason
-
static variable PSQ_DS_GatherAndWriteFrequencyToLabnotebook(string device, variable sweepNo, variable headstage)¶
-
static std::tuple<variable, WAVE, WAVE> PSQ_DS_CalculateMaxSlopeAndWriteToLabnotebook(string device, variable sweepNo, variable headstage, variable fitSlope, variable minimumSpikeCountForMaxSlope)¶
-
static variable PSQ_DS_CalculateReachedFinalSlope(wave fitSlopes, wave DAScales, wave fillinPassed, variable fitSlope, variable maxSlope, variable slopePercentage)¶
Determine the f-I slope QC.
For a passing we need all of the following to be true:
valid fit
finite slope and not PSQ_DS_SKIPPED_FI_SLOPE
slope must be slopePercentage smaller than the maximum slope
must not be from a fillin sweep
slope must be acquired with a larger DAScale value than maximum slope
-
static variable PSQ_DS_CalculateNegativeSlopePass(wave fitSlopes, wave fillinPassed, variable fitSlope)¶
-
static variable PSQ_DS_IsFillin(wave fitSlopes, wave fillinPassed, variable fitSlope)¶
-
static variable PSQ_DS_CalculateFillinQC(wave DAScales, variable dascale)¶
-
static variable PSQ_DS_CalculateReachedFinalSlopeAndWriteToLabnotebook(string device, variable sweepNo, wave fitSlopes, wave DAScales, wave fillinPassed, variable fitSlope, variable maxSlope, variable slopePercentage)¶
-
static variable PSQ_DS_CalculateNegSlopePassAndWriteToLabnotebook(string device, variable sweepNo, wave fitSlopes, wave fillinPassed, variable fitSlope)¶
-
static variable PSQ_DS_CalculateFillinPassAndWriteToLabnotebook(string device, variable sweepNo, wave DAScales)¶
-
static variable PSQ_DS_IsValidFitSlopePosition(wave fitSlopes, wave DAScales, variable fitSlope, variable maxSlope)¶
Check if the given fitSlope is a suitable candidate for maxSlope/negSlope.
The condition is that the fitSlope is not acquired earlier than maxSlope as we want to find the case there it drops off and not where it starts to rise. We do use the DAScales values for this to ensure that measurements from overshoot correction (aka fill-in) don’t have a chance of interfering.
-
static std::tuple<variable, variable> PSQ_DS_StoreFitOffsetAndSlope(string device, variable sweepNo, variable headstage, WaveOrNull fitOffsetAll, string lbnKeyOffset, WaveOrNull fitSlopeAll, string lbnKeySlope)¶
-
static std::tuple<WaveText> PSQ_DS_EvaluateAdaptiveThresholdSweep(string device, variable sweepNo, variable headstage, PSQ_DS_DAScaleParams *cdp, variable minimumSpikeCountForMaxSlope)¶
Evaluate the complete adaptive supra threshold sweep.
Gather AP frequency and DAscale data
Fetch the same data from the passing rheobase/supra sweeps and previous adaptive sweeps from the same SCI
Create a new line fit for the last two points with differing AP frequency
Check if the resulting fit slope is smaller than
slopePercentage
- Return values:
futureDAScales – future DAScale values including the historic ones,
-
static variable PSQ_DS_AdaptiveDetermineSweepQCResults(string device, variable sweepNo, variable headstage, variable numSweepsWithSaturation)¶
Determine from the POST_SWEEP_EVENT if the set is already finished.
We don’t check here for passing/failing set QC but only if we need to continue acquisition.
-
static std::tuple<string, string, variable> PSQ_DS_GetLabnotebookKeyConstants(variable type)¶
Helper function for PSQ_DS_GetLabnotebookData.
- Parameters:
type – One of PSQDAScaleAdaptiveLBNTypeConstants
- Return values:
currentSCI – labnotebook string (PSQ_FMT_XXX or stock entry) for data from the current SCI
RhSuAd – labnotebook PSQ_FMT_XXX string for RhSuAd data
headstageContingencyMode – One of HCM_INDEP/HCM_DEPEND
-
static std::tuple<WAVE, variable> PSQ_DS_GetLabnotebookData(wave numericalValues, wave textualValues, variable sweepNo, variable headstage, variable type, variable filterPassing = defaultValue, variable filterNegSlopeAndNaN = defaultValue, variable beforeSweepQCResult = defaultValue, variable fromRhSuAd = defaultValue)¶
Return some labnotebook entries for all passing sweeps from RhSuAd and the current SCI.
-
static variable PSQ_DS_NegativefISlopePassingCriteria(wave numericalValues, WaveText textualValues, variable sweepNo, variable headstage, variable fromRhSuAd = defaultValue)¶
-
static variable PSQ_DS_AddDAScaleFillinBetweenPosAndNegSlope(string device, variable sweepNo, variable headstage, variable fromRhSuAd = defaultValue)¶
Add fillin value between the last positive f-I slope and the next negative f-I slope.
- Return values:
zero – if a value could be added, one if not
-
static variable PSQ_DS_AdaptiveIsFinished(string device, variable sweepNo, variable headstage, variable numSweepsWithSaturation, variable fromRhSuAd = defaultValue)¶
-
static variable PSQ_DS_ConsecutivePasses(wave refQC, wave checkQC)¶
Return the number of consecutive passing labnotebook entries
checkQCwhich all haverefQCpassing.It is assumed that
checkQCis a value which is calculated for two neighbouring sweeps.
-
static variable PSQ_DS_HasRequiredQCOrder(wave firstQC, wave checkQCFirst, wave lastQC, wave checkQCLast)¶
Searches for exactly one passing check of each checkQCFirst and checkQCLast (in this order) which both have
firstQC/lastQCpassing and only failingfirstQC/lastQCin between.
-
static wave PSQ_DS_GetFutureDAScalesFromLBN(wave numericalValues, wave textualValues, variable sweepNo, variable headstage)¶
-
static std::tuple<WaveText, WAVE, WAVE> PSQ_DS_GatherFutureDAScalesAndFrequency(string device, variable sweepNo, variable headstage, PSQ_DS_DAScaleParams *cdp)¶
-
static variable PSQ_DS_CalcFutureDAScalesAndStoreInLBN(string device, variable sweepNo, variable headstage, WaveText futureDAScales)¶
-
static wave PSQ_DS_FilterPassingData(WaveOrNull data, WaveOrNull booleanQC, variable pairwise = defaultValue, variable inBetween = defaultValue)¶
Return the entries of data which have passing state in booleanQC.
Both data and booleanQC are usually from the full SCI.
- Parameters:
data – wave to filter
booleanQC – used for filter decision
pairwise – [optional, defaults to false] decision wave is one element shorter as it applies pairwise
inBetween – [optional, defaults to flase] decision wave is one element larger (reverse of pairwise)
-
static wave PSQ_DS_GetPassingDAScaleSweeps(wave numericalValues, variable sweepNo, variable headstage)¶
Return a wave with all passing sweeps from the SCI containing sweepNo/headstage.
-
static wave PSQ_DS_GetPassingRheobaseSweeps(wave numericalValues, variable passingRheobaseSweep, variable headstage)¶
-
static wave PSQ_DS_GetPassingRhSuAdSweepsAndStoreInLBN(string device, variable sweepNo, variable headstage, string params)¶
-
static std::tuple<variable, variable> PSQ_DS_CalculateAndStoreDAScaleStepWidths(string device, variable sweepNo, wave apfreqRhSuAd, wave DAScalesRhSuAd, variable maxFrequencyChangePercent, variable dascaleStepWidthMinMaxRatio, variable fallbackDAScaleRangeFac)¶
Calculate the minimum and maximum step widths for DAScale normalized.
Stored in the labnotebook.
Multiply by
AP Frequencyto get scaled values.
-
static std::tuple<variable, variable, variable, variable, variable> PSQ_DS_GetValuesOfLargestDAScale(wave numericalValues, WaveText textualValues, variable sweepNo, variable headstage, variable fromRhSuAd = defaultValue)¶
-
static variable PSQ_DS_CalculateMaxSlope(wave fitSlopes, wave apfreq, variable minimumSpikeCountForMaxSlope)¶
-
static variable PSQ_DS_DAScaleParamsAreFinite(PSQ_DS_DAScaleParams *cdp)¶
-
static variable PSQ_DS_InitDAScaleParams(PSQ_DS_DAScaleParams *cdp)¶
-
static string PSQ_GetHelpCommon(variable type, string name)¶
Help strings for common parameters.
Not every analysis function uses every parameter though.
-
static string PSQ_CheckParamCommon(string name, CheckParametersStruct *s, variable maxThreshold = defaultValue)¶
-
string PSQ_DAScale_GetParams()¶
Require parameters from stimset.
-
string PSQ_DAScale_GetHelp(string name)¶
-
string PSQ_DAScale_CheckParam(string name, CheckParametersStruct *s)¶
-
variable PSQ_DAScale(string device, AnalysisFunction_V3 *s)¶
Patch Seq Analysis function to find a suitable DAScale.
Prerequisites:
Does only work for one headstage
Assumes that the stimset has 500ms of pre pulse baseline, a non-zero pulse and at least 1000ms post pulse baseline.
Each 500ms (PSQ_BL_EVAL_RANGE) of the baseline is a chunk
Testing: For testing the spike detection logic, the results can be defined in the override wave,
Reading the results from the labnotebook:
See also
WAVE numericalValues = GetLBNumericalValues(device) // set properties key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_SET_PASS, query = 1) setPassed = GetLastSettingIndepSCI(numericalValues, sweepNo, key, headstage, UNKNOWN_MODE) if(setPassed) // set passed else // set did not pass endif // single sweep properties key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_SWEEP_PASS, query = 1) sweepPassed = GetLastSettingIndep(numericalValues, sweepNo, key, UNKNOWN_MODE) // chunk (500ms portions of the baseline) properties key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_CHUNK_PASS, chunk = chunk, query = 1) chunkPassed = GetLastSettingIndep(numericalValues, sweepNo, key, UNKNOWN_MODE) // single test properties key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_RMS_SHORT_PASS, chunk = chunk, query = 1) rmsShortPassed = GetLastSettingIndep(numericalValues, sweepNo, key, UNKNOWN_MODE) key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_RMS_LONG_PASS, chunk = chunk, query = 1) rmsLongPassed = GetLastSettingIndep(numericalValues, sweepNo, key, UNKNOWN_MODE) key = CreateAnaFuncLBNKey(PSQ_DA_SCALE, PSQ_FMT_LBN_TARGETV_PASS, chunk = chunk, query = 1) targetVPassed = GetLastSettingIndep(numericalValues, sweepNo, key, UNKNOWN_MODE) // get fitted resistance from last passing sweep // resistance for the first headstage can be found in resistanceFitted[0] WAVE/Z resistanceFitted = GetLastSettingSCI(numericalValues, sweepNo, LABNOTEBOOK_USER_PREFIX + LBN_RESISTANCE_FIT, headstage, UNKNOWN_MODE)
Decision logic flowchart:
/// /// Sketch of a stimset with inserted TP, pre pulse baseline (-), pulse (*), and post pulse baseline (-). /// /// |-| *********************************** /// | | | | /// | | | \WWW/ | /// | | | / \ | /// | | | /wwwww\ | /// | | | _| o_o |_ | /// | | | \WWWWWWW/ (_ / \ _) | /// | | | _/` o_o `\_ | \_/ | | /// | | | (_ (_) _) : ~~~~~ : | /// | | | \ '-...-' / \_____/ | /// | | | (`'-----'`) [ ] | /// | | | `"""""""` `"""""` | /// | | | | /// -| |-----------------| |-------------------------------------------- /// /// ascii art image from: http://ascii.co.uk/art/erniebert /// ///
-
string PSQ_SquarePulse_GetParams()¶
Return a list of required parameters.
-
string PSQ_SquarePulse_GetHelp(string name)¶
-
string PSQ_SquarePulse_CheckParam(string name, CheckParametersStruct *s)¶
-
variable PSQ_SquarePulse(string device, AnalysisFunction_V3 *s)¶
Analysis function to find the smallest DAScale where the cell spikes.
Prerequisites:
Does only work for one headstage
Assumes that the stimset has a pulse
Testing: For testing the spike detection logic, the results can be defined in the override wave,
Decision logic flowchart:
See also
/// /// Sketch of a stimset with pre pulse baseline (-), pulse (*), and post pulse baseline (-). /// /// ******* /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// ---| |-------------------------------------------- /// ///
-
string PSQ_Rheobase_GetParams()¶
Return a list of required parameters.
-
string PSQ_Rheobase_GetHelp(string name)¶
-
string PSQ_Rheobase_CheckParam(string name, CheckParametersStruct *s)¶
-
variable PSQ_Rheobase(string device, AnalysisFunction_V3 *s)¶
Analysis function for finding the exact DAScale value between spiking and non-spiking.
Prerequisites:
Does only work for one headstage
Assumes that the stimset has a pulse of non-zero and arbitrary length
Pre pulse baseline length is PSQ_BL_EVAL_RANGE
Post pulse baseline length a multiple of PSQ_BL_EVAL_RANGE
Testing: For testing the spike detection logic, the results can be defined in the override wave,
Decision logic flowchart:
See also
/// /// Sketch of a stimset with pre pulse baseline (-), pulse (*), and post pulse baseline (-). /// /// ******* /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// | | /// ---| |-------------------------------------------- /// ///
-
variable PSQ_GetFinalDAScaleFake()¶
-
string PSQ_Ramp_GetParams()¶
Return a list of required parameters.
-
string PSQ_Ramp_GetHelp(string name)¶
-
string PSQ_Ramp_CheckParam(string name, CheckParametersStruct *s)¶
-
variable PSQ_Ramp(string device, AnalysisFunction_V3 *s)¶
Analysis function for applying a ramp stim set and finding the position were it spikes.
Prerequisites:
Does only work for one headstage
Assumes that the stimset has a ramp of non-zero and arbitrary length
Pre pulse baseline length is PSQ_BL_EVAL_RANGE
Post pulse baseline length is at least two times PSQ_BL_EVAL_RANGE
Testing: For testing the spike detection logic, the results can be defined in the override wave,
Decision logic flowchart:
See also
/// /// Sketch of a stimset with pre pulse baseline (-), ramp (/|), and post pulse baseline (-). /// /// /| /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// / | /// ---/ |-------------------------------------------- /// ///
-
static variable PSQ_Ramp_AddEpoch(string device, variable headstage, wave wv, string tags, string shortName, variable first, variable last)¶
-
variable PSQ_Ramp_AddEpochImpl(WaveText epochWave, wave wv, variable DAC, string tags, string shortName, variable first, variable last)¶
device independent implementation of PSQ_Ramp_AddEpoch
-
std::tuple<variable, variable> PSQ_CR_SetHasPassed(wave numericalValues, variable sweepNo, variable headstage)¶
Determine if we have three passing sweeps with the same DAScale value.
- Returns:
result set passing state (0/1)
- Returns:
maxOccurences maximum number of passing sets with the same DASCale value
-
static std::tuple<ChirpBoundsInfo> PSQ_CR_DetermineBoundsState(variable baseline, variable minimum, variable maximum, variable value)¶
Returns the two letter states “AA”, “BB” and “BA” for the value and the scaling factors to reach min/center/max.
We need the real baseline value (measured), whereas the center between minimum/maximum would be targetV, which the user entered together with an allowed ± range.
-
string PSQ_CR_BoundsActionToString(variable boundsAction)¶
-
static variable PSQ_CR_DetermineBoundsActionHelper(wave wv, string state, variable action)¶
-
static variable PSQ_CR_DetermineBoundsActionFromState(string upperState, string lowerState)¶
Returns an action depending on the upper and lower states, see ChirpBoundsAction.
-
static variable PSQ_CR_DetermineScalingFactor(ChirpBoundsInfo *lowerInfo, ChirpBoundsInfo *upperInfo, variable boundsEvaluationMode)¶
Determine the scaling factor for DAScale which is required for being inside the bounds.
-
static std::tuple<variable, variable> PSQ_CR_DetermineBoundsAction(string device, wave scaledDACWave, variable headstage, variable sweepNo, variable chirpStart, variable cycleEnd, variable innerRelativeBound, variable outerRelativeBound, variable boundsEvaluationMode)¶
Determine the bounds action given the requested chirp slice.
- Parameters:
device – device
scaledDACWave – DAQ data wave with correct units and scaling
headstage – headstage
sweepNo – sweep number
chirpStart – x-position relative to stimset start where the chirp starts
cycleEnd – x-position relative to stimset start where the requested number of cycles finish
innerRelativeBound – analysis parameter
outerRelativeBound – analysis parameter
boundsEvaluationMode – bounds evaluation mode, one of PSQChirpBoundsEvaluationMode
- Returns:
boundsAction, one of ChirpBoundsAction
- Returns:
scalingFactorDAScale, scaling factor to be inside the bounds for actions PSQ_CR_INCREASE/PSQ_CR_DECREASE
-
static string PSQ_CR_BoundsEvaluationModeToString(variable val)¶
-
static variable PSQ_CR_ParseBoundsEvaluationModeString(string str)¶
-
static variable PSQ_SetAutobiasTargetVIfPresent(string device, variable headstage, string params, string name)¶
-
static variable PSQ_SetAutobiasTargetV(string device, variable headstage, variable value)¶
-
static variable PSQ_CR_SetAutobiasTargetVFromTrueRMP(string device, variable headstage, string params)¶
Set autobias target V from the average voltage of a passing PSQ_TrueRestingMembranePotential() sweep.
- Returns:
0 on success, 1 otherwise
-
string PSQ_Chirp_GetHelp(string name)¶
-
string PSQ_Chirp_CheckParam(string name, CheckParametersStruct *s)¶
-
string PSQ_Chirp_GetParams()¶
Return a list of required analysis functions for PSQ_Chirp()
-
variable PSQ_Chirp(string device, AnalysisFunction_V3 *s)¶
Analysis function for determining the impedance of the cell using a sine chirp stim set.
Prerequisites:
Does only work for one headstage
Assumes that the stimset has a chirp of non-zero and arbitrary length
Pre pulse baseline length is PSQ_BL_EVAL_RANGE
Post pulse baseline length is at least two times PSQ_BL_EVAL_RANGE
Testing: For testing the range detection logic, the results can be defined in the wave root:overrideResults.
Decision logic flowchart:
See also
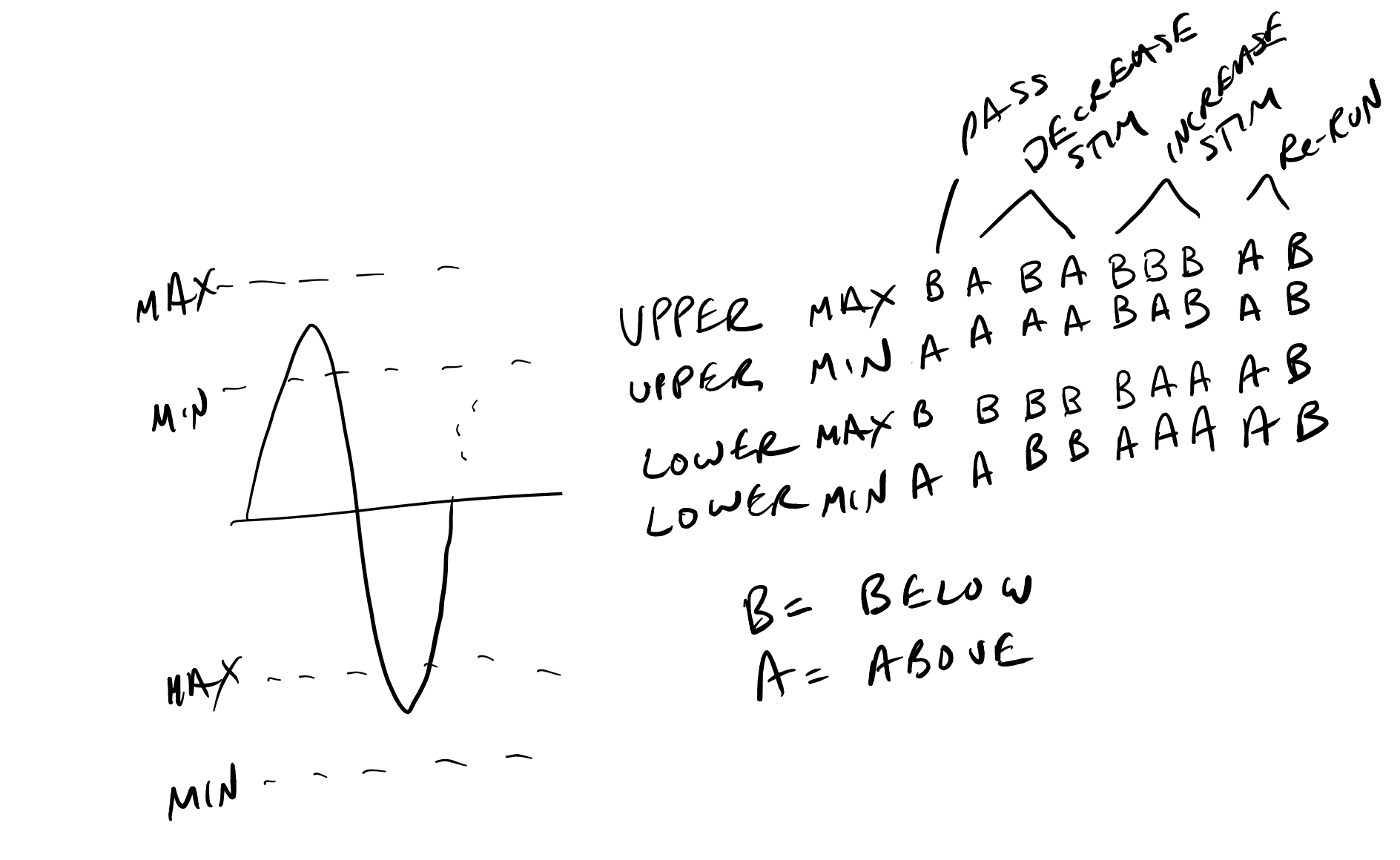
The bounds action for Symmetric (Upper and Lower) is derived from the state according to the following sketch:
Depolarized (Upper only):
State
Action
BA
Pass
AA
Decrease
BB
Increase
Hyperpolarized (Lower only):
State
Action
BA
Pass
AA
Increase
BB
Decrease
/// /// Sketch of a stimset with pre pulse baseline (-), sine chirp (~), and post pulse baseline (-). /// /// /// /// ~ ~ /// ~ ~ ~ ~ /// ~ ~ ~ ~ /// ~ ~ ~ ~ /// ---------------~ ~ ~ ~-------------------------------- /// ~ ~ /// ~ ~ /// ~ /// ///
-
static variable PSQ_CR_FindDependentAnalysisParameter(string device, string name, string params)¶
-
static std::tuple<variable, variable> PSQ_CR_GetSpikeEvaluationRange(string device, variable sweepNo, variable headstage)¶
Return the begin/start [ms] of the spike bounds evaluation range.
Zero is the DA/AD wave zero.
-
std::tuple<variable, variable> PSQ_CR_AddSpikeEvaluationEpoch(WaveText epochsWave, variable DAC, variable headstage, wave durations, variable totalOnsetDelayMS)¶
Adds PSQ_Chirp spike evaluation epoch.
- Parameters:
epochsWave – 4d epochs wave
DAC – DAC number
headstage – headstage number
durations – pulse duration wave
totalOnsetDelayMS – totalOnsetDelay in ms
- Return values:
epBegin – epoch begin in s
epEnd – epoch end in s
-
static std::tuple<variable, variable> PSQ_CR_GetChirpEvaluationRange(string device, variable sweepNo, variable headstage, variable requestedCycles)¶
Return the begin/start [ms] of the chirp bounds evaluation range.
Zero is the DA/AD wave zero.
-
std::tuple<variable, variable> PSQ_CR_AddCycleEvaluationEpoch(WaveText epochsWave, WaveText fullCycleEpochs, variable DAC)¶
Add PSQ_Chirp cycle evaluation epoch.
- Parameters:
epochsWave – 4d epoch wave
fullCycleEpochs – 2d epoch wave with epochs of full cycles (sorted)
DAC – DAC number
- Return values:
epBegin – epoch begin in s
epEnd – epoch end in s
-
wave PSQ_CR_GetFullCycleEpochs(wave numericalValues, WaveText textualValues, variable DAC, WaveText epochsWave, variable requestedCycles)¶
-
static variable PSQ_ForceSetEvent(string device, variable headstage)¶
Manually force the pre/post set events.
Required to do before skipping sweeps.
- Todo:
this hack must go away.
-
static string PSQ_ExecuteSweepFormula(string device, string code)¶
Execute
codein the SweepFormula notebook.
-
static variable PSQ_SetSamplingIntervalMultiplier(string device, variable multiplier)¶
-
static variable PSQ_SetStimulusSets(string device, variable headstage, string params)¶
-
string PSQ_PipetteInBath_CheckParam(string name, CheckParametersStruct *s)¶
-
string PSQ_PipetteInBath_GetHelp(string name)¶
-
string PSQ_PipetteInBath_GetParams()¶
-
variable PSQ_PipetteInBath_GetNumberOfTestpulses(string params)¶
-
variable PSQ_PipetteInBath(string device, AnalysisFunction_V3 *s)¶
Analysis function for determining the pipette resistance while that is located in the bath.
Prerequisites:
Does only work for one headstage
We expect three epochs per test pulse in the stimset, plus one epoch before all TPs, and at the very end
Assumes that the stimset has
NumberOfTestpulsestest pulsesPre pulse baseline length is PSQ_BL_EVAL_RANGE
Post pulse baseline is not evaluated
Testing: For testing the range detection logic, the results can be defined in the wave root:overrideResults.
Decision logic flowchart:
See also
/// /// Sketch of a stimset with three test pulse like epochs /// /// +-----+ +-----+ +-----+ /// | | | | | | /// | | | | | | /// | | | | | | /// -------------------+ +-------+ +-------+ +------------------------ /// /// Epoch borders /// ------------- /// /// 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 /// ///
-
static wave PSQ_GetSweepFormulaResultWave(WaveText textualResultsValues, string key)¶
Return entries from the textual result wave written using
storewith the givenkeyNote: This function always returns the last entry, so this needs to be called directly after PSQ_ExecuteSweepFormula without giving control back to Igor Pro.
-
static variable PSQ_PB_GetPrePulseBaselineDuration(string device, variable headstage)¶
-
static variable PSQ_CreateTestpulseEpochs(string device, variable headstage, variable numTestPulses)¶
Create user epochs for the testpulse like shapes in the stimset.
Assumes that all sweeps in the stimset are the same.
- Returns:
0 on success, 1 on failure
-
variable PSQ_CreateTestpulseEpochsImpl(WaveText epochWave, variable DAC, string setName, variable totalOnsetDelay, variable DAScale, variable numTestPulses)¶
Device independent implementation function for PSQ_CreateTestpulseEpochs.
Assumes that all sweeps in the stimset are the same.
- Returns:
0 on success, 1 on failure
-
static variable PSQ_CreateTestpulseLikeEpoch(WaveText epochWave, variable DAC, string setName, variable DAScale, variable start, variable epochIndex, variable tpIndex)¶
-
string PSQ_SealEvaluation_CheckParam(string name, CheckParametersStruct *s)¶
-
string PSQ_SealEvaluation_GetHelp(string name)¶
-
string PSQ_SealEvaluation_GetParams()¶
-
variable PSQ_SealEvaluation(string device, AnalysisFunction_V3 *s)¶
Analysis function for checking the seal resistance.
Prerequisites:
Does only work for one headstage
See below for the stimset layout
The dedicated baseline epochs before each testpulse group (epoch 0 and 11) and must be at least as large as the given
BaselineChunkLengthanalysis parameter
Testing: For testing the range detection logic, the results can be defined in the wave root:overrideResults.
Decision logic flowchart:
See also
/// /// Sketch of the stimset /// /// +-----+ +-----+ +-----+ +-----+ +-----+ +-----+ /// | | | | | | | | | | | | /// | | | | | | | | | | | | /// | | | | | | | | | | | | /// -------------------+ +-------+ +-------+ +------------------------------------+ +-------+ +-------+ +------------ /// /// Epoch borders /// ------------- /// /// 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12| 13 | 14| 15| 16 | 17|18 | 19 | 20 | 21 | /// /// We always expect PSQ_SE_REQUIRED_EPOCHS (22) epochs even if the user only chose `First` or `Second` on testpulse selection. /// /// BEGIN group A /// 0: baselineQC chunk with length `BaselineChunkLength` or longer before group A /// 1-3: 1. TP /// 4-6: 2. TP /// 7-9: 3. TP /// END group A /// 10: empty space, can be zero points long, but is always present /// BEGIN group B /// 11: baselineQC chunk with length `BaselineChunkLength` or longer before group B /// 12-14: 1. TP /// 15-17: 2. TP /// 18-20: 3. TP /// END group B /// 21: trailing empty space /// ///
-
static variable PSQ_SE_CreateEpochs(string device, variable headstage, string params)¶
Create user epochs.
Assumes that all sweeps in the stimset are the same.
- Returns:
0 on success, 1 on failure
-
variable PSQ_SE_CreateEpochsImpl(WaveText epochWave, variable DAC, variable totalOnsetDelayMS, string setName, variable testpulseGroupSel, variable DAScale, variable numEpochs, variable chunkLength)¶
-
variable PSQ_SE_GetTestpulseGroupSelection(string params)¶
-
static variable PSQ_SE_ParseTestpulseGroupSelection(string str)¶
-
string PSQ_TrueRestingMembranePotential_CheckParam(string name, CheckParametersStruct *s)¶
-
string PSQ_TrueRestingMembranePotential_GetHelp(string name)¶
-
string PSQ_TrueRestingMembranePotential_GetParams()¶
-
variable PSQ_TrueRestingMembranePotential(string device, AnalysisFunction_V3 *s)¶
Analysis function for determining the mean potential of a stimset.
Prerequisites:
Does only work for one headstage
See below for the stimset layout
Testing: For testing the range detection logic, the results can be defined in the wave root:overrideResults.
Decision logic flowchart:
See also
/// /// Sketch of the stimset /// /// -------------------------------------------- /// /// Epoch borders /// ------------- /// /// | 0 | 1 | 2 | /// /// We always expect PSQ_VM_REQUIRED_EPOCHS (3) epochs. /// /// 0: baselineQC chunk with length `BaselineChunkLength` or larger /// 1: empty space, can be zero points long, but is always present /// 2: baselineQC chunk with length `BaselineChunkLength` or larger /// ///
-
static std::tuple<string, string> PSQ_CreateBaselineChunkSelectionStrings(variable index)¶
-
static variable PSQ_CreateBaselineChunkSelectionEpochs(string device, variable headstage, string params, wave epochIndizes, variable numRequiredEpochs = defaultValue)¶
Create baseline selection epochs.
Assumes that all sweeps in the stimset are the same.
- Parameters:
device – device
headstage – headstage
params – analysis function parameters
epochIndizes – indizes of stimset epochs for which we should create BLS epochs
numRequiredEpochs – number of required epochs in the stimset, use
- Returns:
0 on success, 1 on failure
-
variable PSQ_CreateBaselineChunkSelectionEpochs_AddEpochs(WaveText epochWave, variable DAC, variable totalOnsetDelayMS, string setName, wave epochIndizes, variable numEpochs, variable chunkLength)¶
-
static variable PSQ_DetermineSweepQCResults(string device, variable type, variable sweepNo, variable headstage, variable requiredPassesInSet, variable numSweepsFailedAllowed)¶
Check if we need to act on the now available sweep QC state.
- Parameters:
device – device
type – analysis function type
sweepNo – sweep number
headstage – headstage
requiredPassesInSet – number of passes in set which makes the set pass
numSweepsFailedAllowed – number of failures in set which makes the set fail
- Returns:
One of DetermineSweepQCReturns
-
static variable PSQ_VM_EvaluateAverageVoltage(string device, variable sweepNo, variable headstage, string params, variable baselineQCPassed)¶
-
static std::tuple<variable, WAVE> PSQ_VM_CheckForSpikes(string device, variable sweepNo, variable headstage, wave scaledDACWave, variable level)¶
-
static variable PSQ_VM_HandleFailingSpikeQC(string device, AnalysisFunction_V3 *s, wave spikePositions)¶
-
static variable PSQ_CheckThatAlarmIsEnabled(string device, wave asyncChannels)¶
-
static variable PSQ_CheckAsyncAlarmStateAndStoreInLabnotebook(string device, variable type, variable sweepNo, wave asyncChannels)¶
-
string PSQ_AccessResistanceSmoke_CheckParam(string name, CheckParametersStruct *s)¶
-
string PSQ_AccessResistanceSmoke_GetHelp(string name)¶
-
string PSQ_AccessResistanceSmoke_GetParams()¶
-
variable PSQ_AccessResistanceSmoke_GetNumberOfTestpulses(string params)¶
-
variable PSQ_AccessResistanceSmoke(string device, AnalysisFunction_V3 *s)¶
Analysis function for determining the TP resistances and their ratio.
Prerequisites:
Does only work for one headstage
We expect three epochs per test pulse in the stimset, plus one epoch before all TPs, and at the very end
Assumes that the stimset has
NumberOfTestpulsestest pulsesPre pulse baseline length is given by
BaselineChunkLengthPost pulse baseline is not evaluated
Testing: For testing the range detection logic, the results can be defined in the wave root:overrideResults.
Decision logic flowchart:
See also
/// /// Sketch of a stimset with three test pulse like epochs /// /// +-----+ +-----+ +-----+ /// | | | | | | /// | | | | | | /// | | | | | | /// -------------------+ +-------+ +-------+ +------------------------ /// /// Epoch borders /// ------------- /// /// 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 /// /// So for this stimset we have two epochs at the very beginning and end, plus /// three epochs per test pulse and three test pulses, which gives eleven in /// total. /// ///
Variables
-
static const double PSQ_BL_PRE_PULSE = 0x0¶
-
static const double PSQ_BL_POST_PULSE = 0x1¶
-
static const double PSQ_BL_GENERIC = 0x2¶
-
static const double PSQ_RMS_SHORT_TEST = 0x0¶
-
static const double PSQ_RMS_LONG_TEST = 0x1¶
-
static const double PSQ_TARGETV_TEST = 0x2¶
-
static const double PSQ_LEAKCUR_TEST = 0x3¶
-
static const double PSQ_DEFAULT_SAMPLING_MULTIPLIER = 4¶
-
static const double PSQ_RHEOBASE_DURATION = 500¶
-
static const double PSQ_DA_FALLBACK_DASCALE_RANGE_FAC = 1.5¶
-
static const double PSQ_DA_FALLBACK_DASCALE_NEG_SLOPE_PERC = 50¶
-
static const double PSQ_DA_FALLBACK_MINIMUM_SPIKES_FOR_MAX_SLOPE = 4¶
-
static const string PSQ_DA_AT_SLOPE_UNIT = "% of Hz/pA"¶
-
struct PSQ_DS_DAScaleParams¶