HMBA Basal Ganglia: Neuroglancer Views#
Neuroglancer Views – Coordinate Spaces#
Click thumbnails to launch Neuroglancer for each coordinate space.
Species |
Donor |
Slab |
Donor |
CCF |
HiP-CT |
|---|---|---|---|---|---|
Human |
H22.30.001 |
|
— |
|

|
Macaque |
QM23.50.001 (Red) |
|
ETA 2026 |
ETA 2026 |
TBD |
Marmoset |
CJ23.56.004 (Tank) |
|
ETA 2026 |
ETA 2026 |
TBD |
The cell coordinates are registered to four different reference spaces, each serving a distinct purpose.
Slab#
The coordinate system of the slab-face images that each spatial transcriptomics plane belongs to.
This view shows cells in the context of the imaging assets used for mosaicking (e.g., block-face and slab-face images).
Donor#
The native MRI space corresponding to the individual donor from whom the spatial transcriptomics sections were sampled.
This view preserves subject-specific anatomy before normalization to any common template.
CCF (Common Coordinate Framework)#
The standardized reference space for the donor’s species, enabling cross-donor and cross-dataset comparisons.
See this link for more details.
HiP-CT (Cellular-Resolution Volumetric Scan)#
A cellular-resolution (25 µm) volumetric scan of the human brain.
Details about the HiP-CT dataset and donor.
Note: The spatial transcriptomics data were not sampled from the same donor; alignment is used only for anatomical correspondence.
Quick Guide#
Click a section title to expand/collapse.
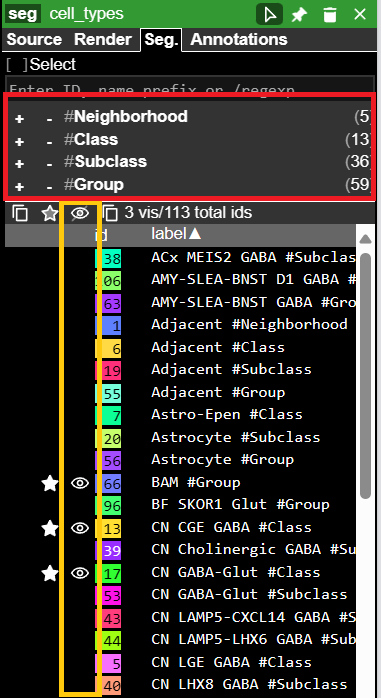
1 · Filter annotations by cell types
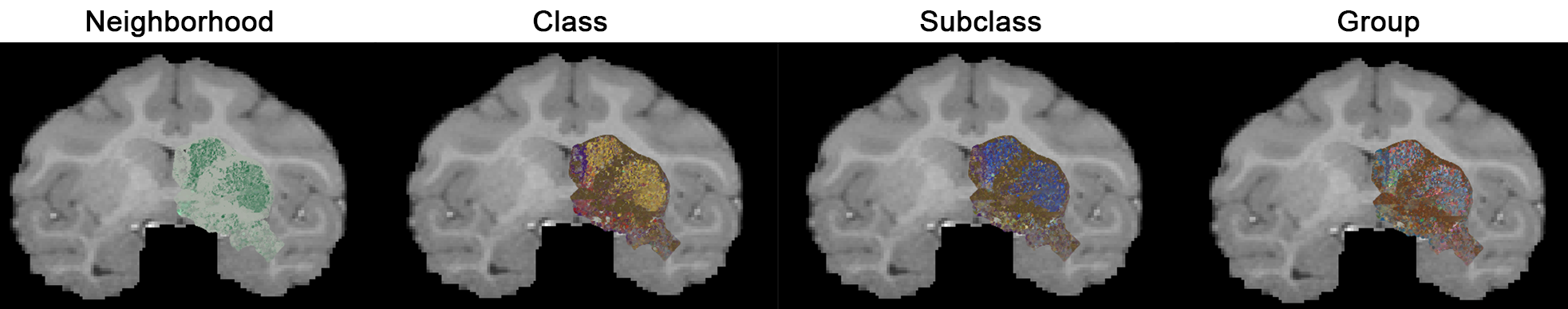
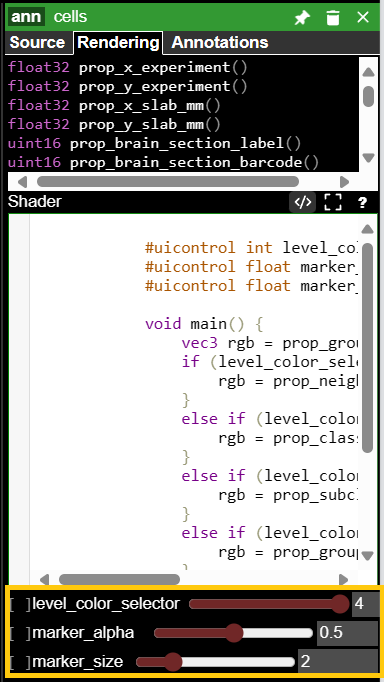
2 · Color annotations by taxonomic level – set annotation size & opacity
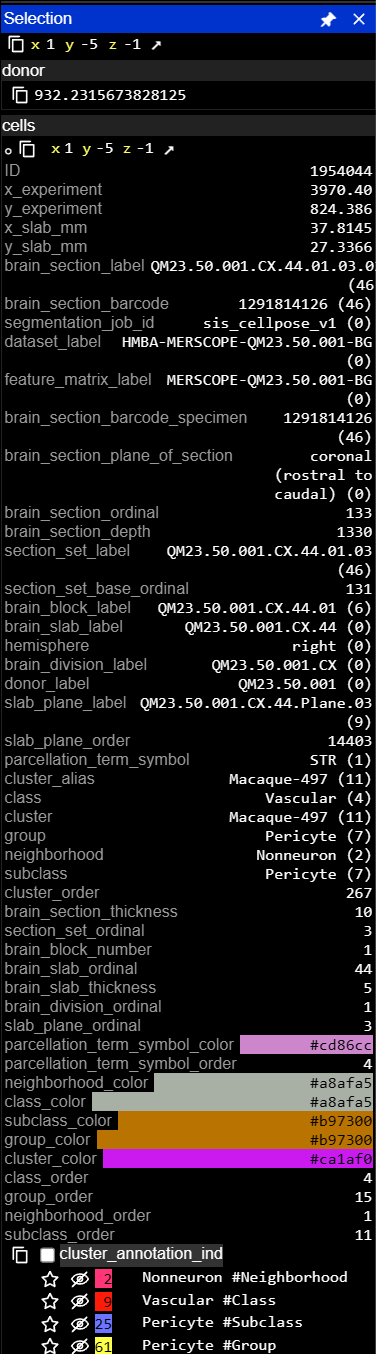
3 · View annotation metadata
4 · Align view with cell sections by tilting cross-section plane
Tilting the cross-section plane helps match views to cell section planes.
Click inside a view (coronal, transverse, sagittal) to focus it.
Rotate with keys: e (left) and r (right).
Or hold Shift and drag with the mouse to tilt across all axes.
Before tilting
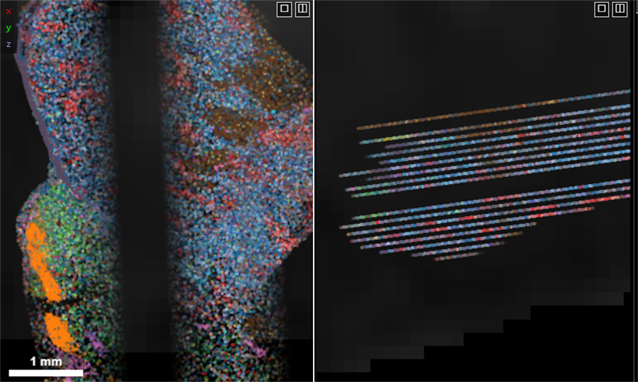
After tilting

5 · Increase annotation view depth
If only a partial coronal cell-section plane is visible, increase the annotation view depth.
Shortcut: Hold Alt and use the mouse wheel.