Visualizing Neuronal Unit Responses#
Note: Some of this content is adapted from the Allen SDK Documentation.
After processing Neuropixels ecephys data with Kilosort, individual neuronal units have been identified and are stored in the units table, in the Units section of the NWB file. The units table contains information about hypothetical neurons determined by Kilosort. With this information and the stimulus information found in the stimulus tables of the Intervals section, this notebook helps examine the spiking behavior of these units in response to stimuli and their associated waveforms
Environment Setup#
⚠️Note: If running on a new environment, run this cell once and then restart the kernel⚠️
import warnings
warnings.filterwarnings('ignore')
try:
from databook_utils.dandi_utils import dandi_download_open
except:
!git clone https://github.com/AllenInstitute/openscope_databook.git
%cd openscope_databook
%pip install -e .
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
Downloading Ecephys File#
Change the values below to download the file you’re interested in. Set dandiset_id and dandi_filepath to correspond to the dandiset id and filepath of the file you want. If you’re accessing an embargoed dataset, set dandi_api_key to your DANDI API key. If you want to stream a file instead of downloading it, use dandi_stream_open instead. Checkout Streaming an NWB File with remfile for more details on this.
dandiset_id = "000021"
dandi_filepath = "sub-703279277/sub-703279277_ses-719161530.nwb"
download_loc = "."
dandi_api_key = None
# This can sometimes take a while depending on the size of the file
io = dandi_download_open(dandiset_id, dandi_filepath, download_loc, dandi_api_key=dandi_api_key)
nwb = io.read()
A newer version (0.62.1) of dandi/dandi-cli is available. You are using 0.55.1
File already exists
Opening file
Extracting Unit Data and Stimulus Data#
Below, the Units table is read from the file. Individual units (putative neurons) are identified with the id column. Note that, while each id is unique, they are not perfectly ordinal; some ids are missing. In the cells below, the unit properties are listed and a slice of the units table is shown. More thorough descriptions of units and their properties can be found in Visualizing Unit Quality Metrics
Additionally, the stimulus data is also read from the NWB file’s Intervals section. Stimulus information is stored as a series of tables depending on the type of stimulus shown in the session. One such table is displayed below.
units = nwb.units
units.colnames
('waveform_duration',
'cluster_id',
'peak_channel_id',
'cumulative_drift',
'amplitude_cutoff',
'snr',
'recovery_slope',
'isolation_distance',
'nn_miss_rate',
'silhouette_score',
'velocity_above',
'quality',
'PT_ratio',
'l_ratio',
'velocity_below',
'max_drift',
'isi_violations',
'firing_rate',
'amplitude',
'local_index',
'spread',
'waveform_halfwidth',
'd_prime',
'presence_ratio',
'repolarization_slope',
'nn_hit_rate',
'spike_times',
'spike_amplitudes',
'waveform_mean')
units[:10]
| waveform_duration | cluster_id | peak_channel_id | cumulative_drift | amplitude_cutoff | snr | recovery_slope | isolation_distance | nn_miss_rate | silhouette_score | ... | local_index | spread | waveform_halfwidth | d_prime | presence_ratio | repolarization_slope | nn_hit_rate | spike_times | spike_amplitudes | waveform_mean | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||
| 950921187 | 0.604355 | 4 | 850249267 | 481.80 | 0.425574 | 2.209140 | -0.118430 | 17.537571 | 0.009496 | 0.036369 | ... | 4 | 50.0 | 0.357119 | 2.962274 | 0.99 | 0.381716 | 0.473829 | [1.0439430431793884, 1.543311060144649, 2.7287... | [0.0001908626967721937, 0.00016134635752077775... | [[0.0, 0.5961149999999966, 5.378099999999993, ... |
| 950921172 | 0.521943 | 3 | 850249267 | 681.53 | 0.390098 | 1.959983 | -0.109729 | 14.677643 | 0.003857 | 0.103446 | ... | 3 | 40.0 | 0.260972 | 2.067810 | 0.99 | 0.536663 | 0.445946 | [10.406435026164546, 17.127986534673788, 18.48... | [0.00014485615850768024, 0.0001722424107984555... | [[0.0, -1.341600000000002, -0.4586399999999933... |
| 950921152 | 0.467002 | 2 | 850249267 | 1070.71 | 0.500000 | 2.522905 | -0.109867 | 15.783665 | 0.017776 | 0.027818 | ... | 2 | 50.0 | 0.247236 | 2.220043 | 0.99 | 0.566559 | 0.284058 | [1.2775103414155262, 2.3915133536963493, 3.701... | [0.00014859435856024575, 0.0001531048673600236... | [[0.0, -0.6427199999999993, -2.836079999999998... |
| 950921135 | 0.467002 | 1 | 850249267 | 253.42 | 0.500000 | 2.803475 | -0.150379 | 26.666930 | 0.023742 | 0.076530 | ... | 1 | 40.0 | 0.233501 | 2.339206 | 0.99 | 0.669090 | 0.590737 | [9.473732504122962, 13.198542576065163, 18.302... | [0.00032386170367170055, 0.0004518112387675137... | [[0.0, -3.2800950000000078, -6.087510000000009... |
| 950921111 | 0.439531 | 0 | 850249267 | 141.82 | 0.018056 | 4.647943 | -0.328727 | 66.901065 | 0.006595 | NaN | ... | 0 | 30.0 | 0.219765 | 4.395994 | 0.99 | 1.261416 | 0.952667 | [1.1677100445138795, 1.1707767194728813, 1.349... | [0.00015644521007973124, 0.000214412247939483,... | [[0.0, -0.9291749999999945, -6.120270000000007... |
| 950927711 | 1.455946 | 482 | 850249273 | 2.46 | 0.000895 | 1.651500 | -0.039932 | 39.400278 | 0.000033 | NaN | ... | 464 | 100.0 | 0.274707 | 5.557479 | 0.37 | 0.467365 | 0.000000 | [2613.8652081509977, 2624.5193369599215, 2734.... | [0.00012946663895843286, 0.0001203425053985725... | [[0.0, 6.3216435986159105, 10.324204152249129,... |
| 950921285 | 2.087772 | 11 | 850249273 | 318.53 | 0.036848 | 1.379817 | NaN | 27.472722 | 0.000903 | 0.291953 | ... | 11 | 100.0 | 0.288442 | 2.751337 | 0.89 | 0.372116 | 0.258065 | [39.04904580954626, 39.457346913598556, 40.495... | [7.768399792002802e-05, 8.405736507197006e-05,... | [[0.0, 5.330324999999991, 2.4261899999999486, ... |
| 950921271 | 0.947739 | 10 | 850249273 | 1008.50 | 0.001727 | 1.420617 | -0.008204 | 30.027595 | 0.000707 | 0.406673 | ... | 10 | 100.0 | 0.288442 | 3.847234 | 0.96 | 0.498618 | 0.796491 | [16.751918851114475, 26.127977537450867, 28.65... | [0.00016516929470324686, 0.0001501058102103845... | [[0.0, -3.103230000000032, 5.680349999999983, ... |
| 950921260 | 0.453266 | 9 | 850249273 | 175.00 | 0.000081 | 4.969091 | -0.184456 | 89.804006 | 0.000000 | 0.223876 | ... | 9 | 60.0 | 0.192295 | 5.274090 | 0.99 | 1.140487 | 0.997333 | [0.9620761551434307, 2.092045877265143, 2.4040... | [0.0003836112198262231, 0.0004093908262843732,... | [[0.0, 1.9104149999999982, -7.270770000000016,... |
| 950921248 | 0.439531 | 8 | 850249273 | 261.11 | 0.065478 | 2.147758 | -0.085677 | 84.145512 | 0.005474 | 0.076346 | ... | 8 | 60.0 | 0.274707 | 3.022387 | 0.99 | 0.463570 | 0.976667 | [1.316477113448928, 1.779311698293908, 2.96088... | [0.00013780619334124896, 0.0001439873831056905... | [[0.0, -3.6761399999999953, 0.8334300000000014... |
10 rows × 29 columns
stimulus_names = nwb.intervals.keys()
print(stimulus_names)
dict_keys(['drifting_gratings_presentations', 'flashes_presentations', 'gabors_presentations', 'invalid_times', 'natural_movie_one_presentations', 'natural_movie_three_presentations', 'natural_scenes_presentations', 'spontaneous_presentations', 'static_gratings_presentations'])
stim_table = nwb.intervals["flashes_presentations"]
stim_table[:]
| start_time | stop_time | stimulus_name | stimulus_block | color | mask | opacity | phase | size | units | stimulus_index | orientation | spatial_frequency | contrast | tags | timeseries | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 0 | 1290.883097 | 1291.133309 | flashes | 1.0 | -1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3647, 1, timestamps pynwb.base.TimeSeries at... |
| 1 | 1292.884817 | 1293.135016 | flashes | 1.0 | 1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3648, 1, timestamps pynwb.base.TimeSeries at... |
| 2 | 1294.886487 | 1295.136691 | flashes | 1.0 | 1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3649, 1, timestamps pynwb.base.TimeSeries at... |
| 3 | 1296.888137 | 1297.138344 | flashes | 1.0 | 1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3650, 1, timestamps pynwb.base.TimeSeries at... |
| 4 | 1298.889787 | 1299.140004 | flashes | 1.0 | -1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3651, 1, timestamps pynwb.base.TimeSeries at... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 145 | 1581.125517 | 1581.375726 | flashes | 1.0 | 1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3792, 1, timestamps pynwb.base.TimeSeries at... |
| 146 | 1583.127217 | 1583.377419 | flashes | 1.0 | 1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3793, 1, timestamps pynwb.base.TimeSeries at... |
| 147 | 1585.128807 | 1585.379029 | flashes | 1.0 | -1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3794, 1, timestamps pynwb.base.TimeSeries at... |
| 148 | 1587.130507 | 1587.380724 | flashes | 1.0 | -1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3795, 1, timestamps pynwb.base.TimeSeries at... |
| 149 | 1589.132187 | 1589.382401 | flashes | 1.0 | -1.0 | None | 1.0 | [0.0, 0.0] | [300.0, 300.0] | deg | 1.0 | 0.0 | [0.0, 0.0] | 0.8 | [stimulus_time_interval] | [(3796, 1, timestamps pynwb.base.TimeSeries at... |
150 rows × 16 columns
Getting Stimulus Epochs#
Here, epochs are extracted from the stimulus tables. In this case, an epoch is a continuous period of time during a session where a particular type of stimulus is shown. The output here is a list of epochs, where an epoch is a tuple of four values; the stimulus name, the stimulus block, the starting time and the ending time. Since stimulus information can vary significantly between experiments and NWB files, you may need to tailor the code below to extract epochs for the file you’re interested in.
# extract epoch times from stim table where stimulus rows have a different 'block' than following row
# returns list of epochs, where an epoch is of the form (stimulus name, stimulus block, start time, stop time)
def extract_epochs(stim_name, stim_table, epochs):
# specify a current epoch stop and start time
epoch_start = stim_table.start_time[0]
epoch_stop = stim_table.stop_time[0]
# for each row, try to extend current epoch stop_time
for i in range(len(stim_table)):
this_block = stim_table.stimulus_block[i]
# if end of table, end the current epoch
if i+1 >= len(stim_table):
epochs.append((stim_name, this_block, epoch_start, epoch_stop))
break
next_block = stim_table.stimulus_block[i+1]
# if next row is the same stim block, push back epoch_stop time
if next_block == this_block:
epoch_stop = stim_table.stop_time[i+1]
# otherwise, end the current epoch, start new epoch
else:
epochs.append((stim_name, this_block, epoch_start, epoch_stop))
epoch_start = stim_table.start_time[i+1]
epoch_stop = stim_table.stop_time[i+1]
return epochs
# extract epochs from all valid stimulus tables
epochs = []
for stim_name in stimulus_names:
stim_table = nwb.intervals[stim_name]
try:
epochs = extract_epochs(stim_name, stim_table, epochs)
except:
continue
# epochs take the form (stimulus name, stimulus block, start time, stop time)
print(len(epochs))
epochs.sort(key=lambda x: x[2])
for epoch in epochs:
print(epoch)
15
('gabors_presentations', 0.0, 89.8968273815905, 1001.8917716749913)
('flashes_presentations', 1.0, 1290.8830973815907, 1589.382401312724)
('drifting_gratings_presentations', 2.0, 1591.1338573815906, 2190.634543106125)
('natural_movie_three_presentations', 3.0, 2221.6604473815905, 2822.161967381591)
('natural_movie_one_presentations', 4.0, 2852.1870373815905, 3152.4377773815904)
('drifting_gratings_presentations', 5.0, 3182.4628573815908, 3781.963503106125)
('natural_movie_three_presentations', 6.0, 4083.215117381591, 4683.716567381592)
('drifting_gratings_presentations', 7.0, 4713.741627381592, 5397.312443106124)
('static_gratings_presentations', 8.0, 5398.31325738159, 5878.714467381591)
('natural_scenes_presentations', 9.0, 5908.739537381591, 6389.157337381589)
('natural_scenes_presentations', 10.0, 6689.408117381589, 7169.809297381591)
('static_gratings_presentations', 11.0, 7199.83431738159, 7680.268867381591)
('natural_movie_one_presentations', 12.0, 7710.293937381591, 8010.54467738159)
('natural_scenes_presentations', 13.0, 8040.56972738159, 8568.510620243858)
('static_gratings_presentations', 14.0, 8611.04613738159, 9151.49746738159)
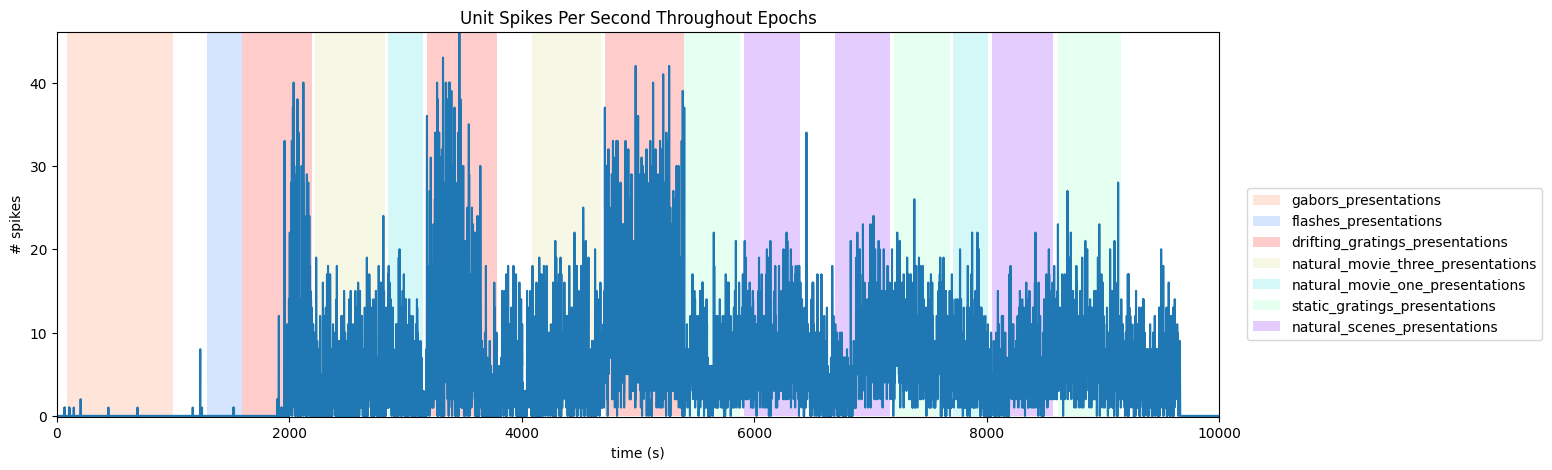
Visualizing Unit Activity Throughout Epochs#
Below is a view of the spiking activity of a unit throughout a session, where epochs are shown as colored sections. Set unit_num to be the id of the unit to view. Set time_start to the starting bound in seconds of the session, you’d like to see, and time_end to the ending bound. You may want to use the output above to inform your choice. As mentioned above, if your file’s stimulus information differs significantly, this code may need to be modified to appropriately display the epochs.
unit_num = 950930672 # chosen from units table
time_start = 0
time_end = 10000
# translate unit id to row index
unit_idx = -1
for i in range(len(units.id)):
if units.id[i] == unit_num:
unit_idx = i
break
print("Unit index:",unit_idx)
Unit index: 648
# make histogram of unit spikes per second over specified timeframe
spikes = units["spike_times"][unit_idx]
time_bin_edges = np.linspace(time_start, time_end, (time_end-time_start))
hist, bins = np.histogram(spikes, bins=time_bin_edges)
# generate plot of spike histogram with colored epoch intervals and legend
fig, ax = plt.subplots(figsize=(15,5))
# assign unique color to each stimulus name
stim_names = list({epoch[0] for epoch in epochs})
colors = plt.cm.rainbow(np.linspace(0,1,len(stim_names)))
stim_color_map = {stim_names[i]:colors[i] for i in range(len(stim_names))}
epoch_key = {}
height = max(hist)
# draw colored rectangles for each epoch
for epoch in epochs:
stim_name, stim_block, epoch_start, epoch_end = epoch
color = stim_color_map[stim_name]
rec = ax.add_patch(mpl.patches.Rectangle((epoch_start, 0), epoch_end-epoch_start, height, alpha=0.2, facecolor=color))
epoch_key[stim_name] = rec
ax.set_xlim(time_start, time_end)
ax.set_ylim(-0.1, height+0.1)
ax.set_xlabel("time (s)")
ax.set_ylabel("# spikes")
ax.set_title("Unit Spikes Per Second Throughout Epochs")
fig.legend(epoch_key.values(), epoch_key.keys(), loc="lower right", bbox_to_anchor=(1.12, 0.25))
ax.plot(bins[:-1], hist)
[<matplotlib.lines.Line2D at 0x116a86b6280>]
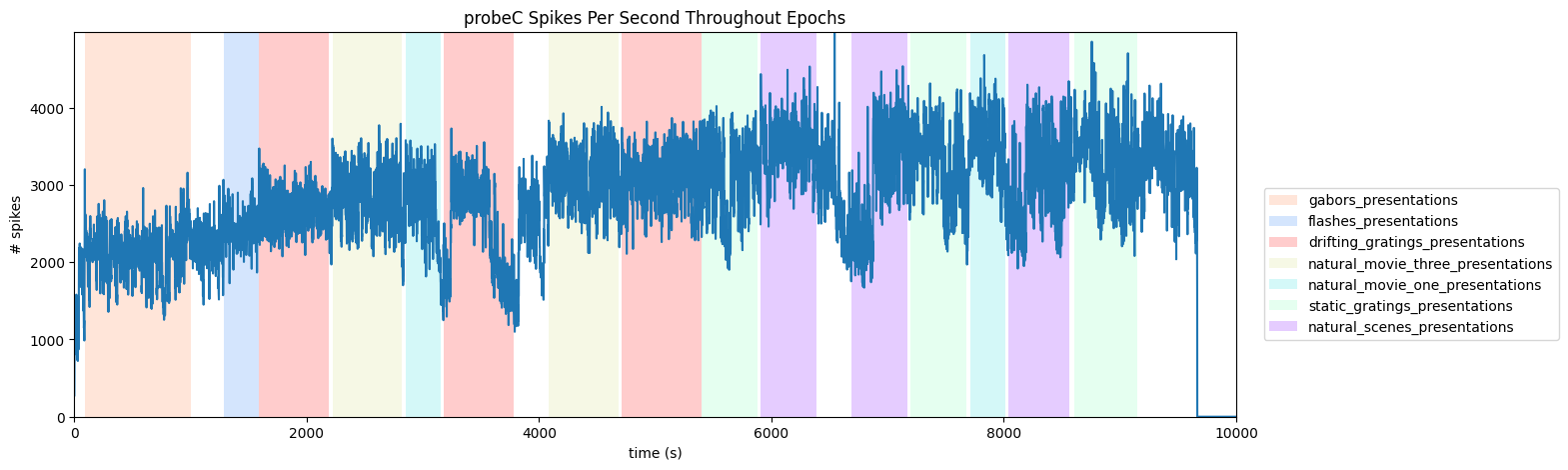
Probewise Activity Throughout Epochs#
It can also be useful to view the activity of an entire probe throughout epochs. The code below allows users to select a probe and a histogram is produced for the total of all unit spiking counts over time for one probe. To do this, the file’s Electrodes table is used. To understand this, you need not know anything about the Electrodes table, except that it can be used to map channel IDs to probe IDs. Below is printed a list of the probe names to choose from. Set probe_name to one of these. Set time_start and time_end to the start and ending times, in seconds, to view in the session.
print(nwb.devices.keys())
dict_keys(['probeA', 'probeB', 'probeC', 'probeD', 'probeE', 'probeF'])
probe_name = "probeC"
time_start = 0
time_end = 10000
# get list of channels on this probe
electrodes = nwb.electrodes.to_dataframe()
channel_ids = set(electrodes.index[electrodes["group_name"] == probe_name].tolist())
# get all spike times from units whose peak channel belongs to the selected probe
probe_spike_times = []
for i in range(len(units)):
if units["peak_channel_id"][i] in channel_ids:
probe_spike_times += list(units["spike_times"][i])
len(probe_spike_times)
27741944
# make histogram of unit spikes per second over specified timeframe
time_bin_edges = np.linspace(time_start, time_end, (time_end-time_start))
hist, bins = np.histogram(probe_spike_times, bins=time_bin_edges)
# generate plot of spike histogram with colored epoch intervals and legend
fig, ax = plt.subplots(figsize=(15,5))
# assign unique color to each stimulus name
stim_names = list({epoch[0] for epoch in epochs})
colors = plt.cm.rainbow(np.linspace(0,1,len(stim_names)))
stim_color_map = {stim_names[i]:colors[i] for i in range(len(stim_names))}
epoch_key = {}
height = max(hist)
# draw colored rectangles for each epoch
for epoch in epochs:
stim_name, stim_block, epoch_start, epoch_end = epoch
color = stim_color_map[stim_name]
rec = ax.add_patch(mpl.patches.Rectangle((epoch_start, 0), epoch_end-epoch_start, height, alpha=0.2, facecolor=color))
epoch_key[stim_name] = rec
ax.set_xlim(time_start, time_end)
ax.set_ylim(-0.1, height+0.1)
ax.set_xlabel("time (s)")
ax.set_ylabel("# spikes")
ax.set_title(f"{probe_name} Spikes Per Second Throughout Epochs")
fig.legend(epoch_key.values(), epoch_key.keys(), loc="lower right", bbox_to_anchor=(1.12, 0.25))
ax.plot(bins[:-1], hist)
[<matplotlib.lines.Line2D at 0x1171e7db820>]
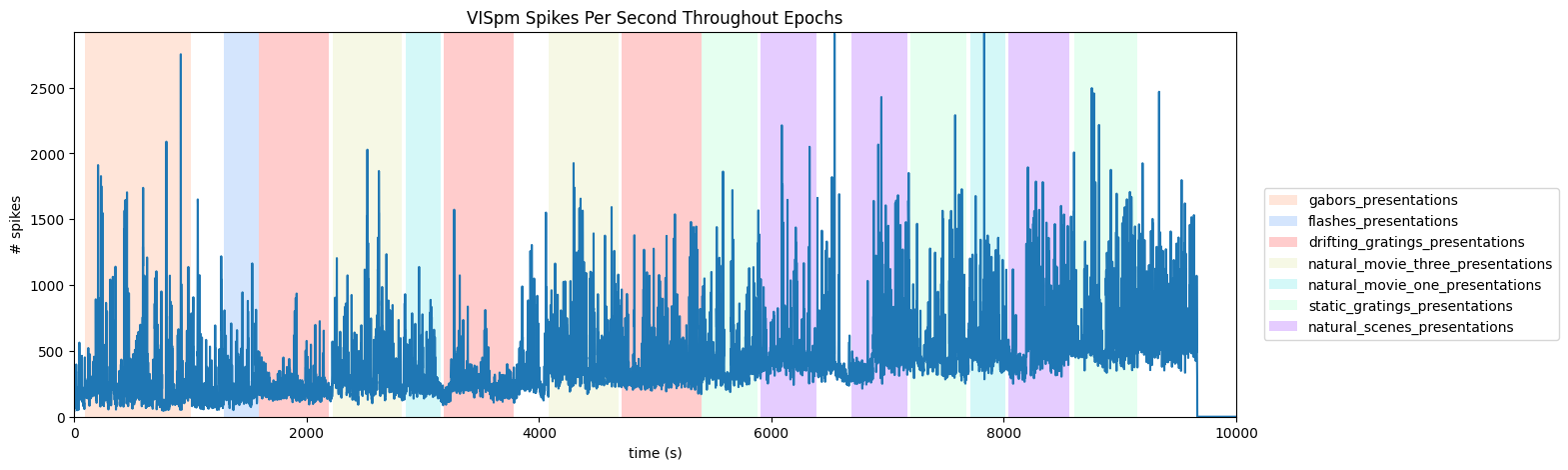
Regionwise Activity Throughout Epochs#
We can also break down our activity based on brain regions (with a bit of work). To do this, we must first be able to retrieve the brain region of each unit. The trick to do this lay in the Electrodes table, shown below. The Electrodes table contains the brain region for each electrode id, while the Units table (shown above) contains the Peak Channel ID for each Unit. These can be used together to get the brain region of each unit’s peak channel. Once this is done, this information can be used just like the Probe selection above to get to get spike counts over time of each Unit in a selected region. Below, set brain_region, start_time, and end_time to view such a plot.
nwb.electrodes[:10]
| x | y | z | imp | location | filtering | group | group_name | probe_vertical_position | probe_horizontal_position | probe_id | local_index | valid_data | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 850249265 | 7955.0 | 3766.0 | 3766.0 | NaN | Eth | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 20 | 43 | 729445650 | 0 | False |
| 850249267 | 7955.0 | 3756.0 | 3756.0 | NaN | TH | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 20 | 11 | 729445650 | 1 | True |
| 850249273 | 7955.0 | 3727.0 | 3727.0 | NaN | TH | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 60 | 43 | 729445650 | 4 | True |
| 850249277 | 7954.0 | 3708.0 | 3708.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 80 | 59 | 729445650 | 6 | True |
| 850249283 | 7954.0 | 3679.0 | 3679.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 100 | 11 | 729445650 | 9 | True |
| 850249289 | 7953.0 | 3640.0 | 3640.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 140 | 43 | 729445650 | 12 | True |
| 850249295 | 7952.0 | 3612.0 | 3612.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 160 | 27 | 729445650 | 15 | True |
| 850249299 | 7952.0 | 3593.0 | 3593.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 180 | 11 | 729445650 | 17 | True |
| 850249305 | 7951.0 | 3565.0 | 3565.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 220 | 43 | 729445650 | 20 | True |
| 850249311 | 7950.0 | 3537.0 | 3537.0 | NaN | APN | AP band: 500 Hz high-pass; LFP band: 1000 Hz l... | probeB abc.EcephysElectrodeGroup at 0x11968190... | probeB | 240 | 27 | 729445650 | 23 | True |
# map electrode ids to brain region acronyms
electrode_locations = {nwb.electrodes["id"][i]: nwb.electrodes["location"][i] for i in range(len(nwb.electrodes))}
print(set(electrode_locations.values()))
{'', 'Eth', 'VISl', 'VPM', 'POL', 'CA1', 'NOT', 'VISam', 'LGd', 'DG', 'VISal', 'APN', 'VISrl', 'SUB', 'VISpm', 'grey', 'PO', 'CA3', 'VISp', 'LP', 'CA2', 'MB', 'TH', 'VL'}
brain_region = "VISpm"
time_start = 0
time_end = 10000
# get all spike times from units whose peak channel belongs to the selected brain region
region_spike_times = []
for i in range(len(units)):
unit_channel = units["peak_channel_id"][i]
unit_location = electrode_locations.get(unit_channel, None)
if unit_location == brain_region:
region_spike_times += list(units["spike_times"][i])
len(region_spike_times)
4178172
# make histogram of unit spikes per second over specified timeframe
time_bin_edges = np.linspace(time_start, time_end, (time_end-time_start))
hist, bins = np.histogram(region_spike_times, bins=time_bin_edges)
# generate plot of spike histogram with colored epoch intervals and legend
fig, ax = plt.subplots(figsize=(15,5))
# assign unique color to each stimulus name
stim_names = list({epoch[0] for epoch in epochs})
colors = plt.cm.rainbow(np.linspace(0,1,len(stim_names)))
stim_color_map = {stim_names[i]:colors[i] for i in range(len(stim_names))}
epoch_key = {}
height = max(hist)
# draw colored rectangles for each epoch
for epoch in epochs:
stim_name, stim_block, epoch_start, epoch_end = epoch
color = stim_color_map[stim_name]
rec = ax.add_patch(mpl.patches.Rectangle((epoch_start, 0), epoch_end-epoch_start, height, alpha=0.2, facecolor=color))
epoch_key[stim_name] = rec
ax.set_xlim(time_start, time_end)
ax.set_ylim(-0.1, height+0.1)
ax.set_xlabel("time (s)")
ax.set_ylabel("# spikes")
ax.set_title(f"{brain_region} Spikes Per Second Throughout Epochs")
fig.legend(epoch_key.values(), epoch_key.keys(), loc="lower right", bbox_to_anchor=(1.12, 0.25))
ax.plot(bins[:-1], hist)
[<matplotlib.lines.Line2D at 0x1173a0d3910>]
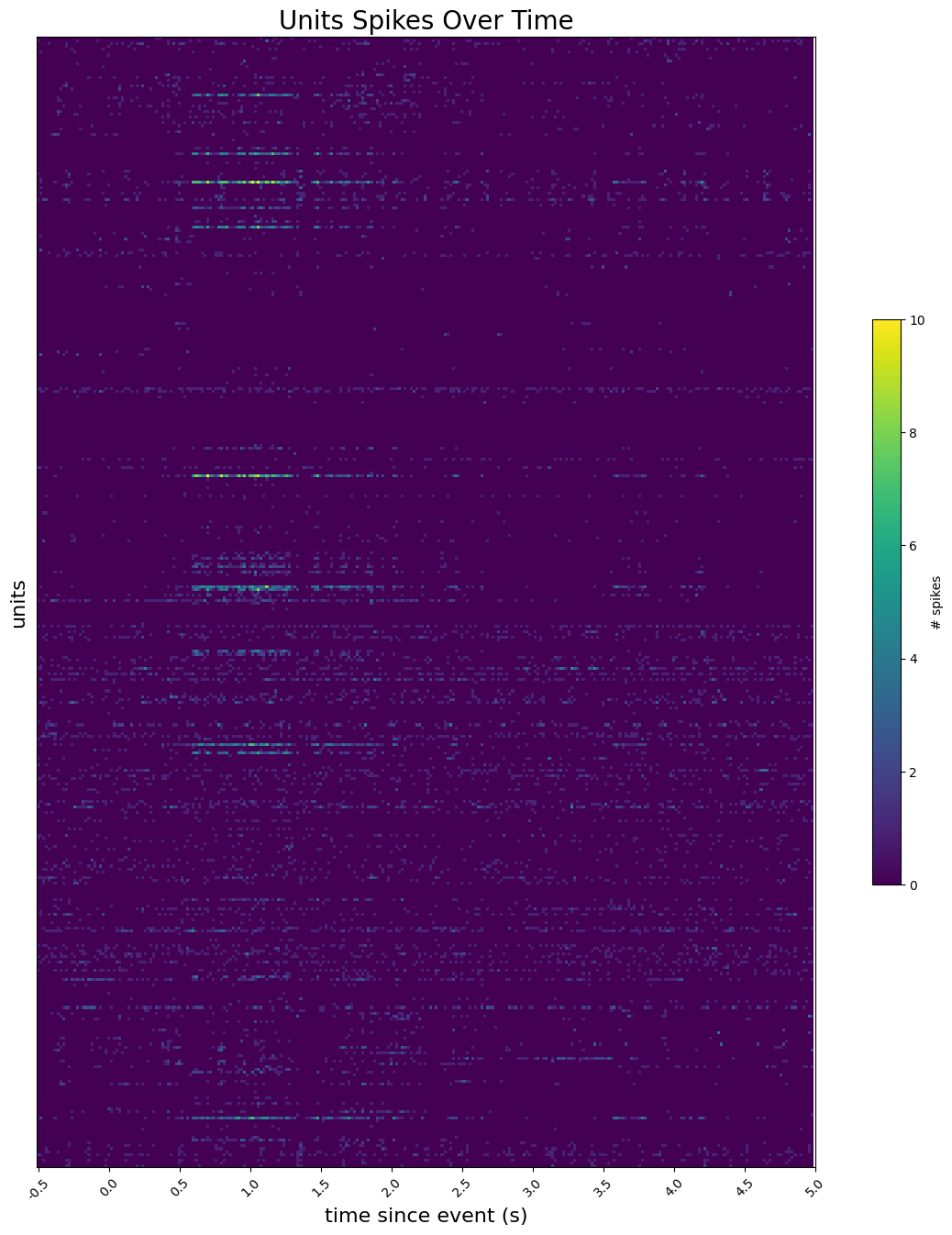
Showing Spike Times#
Here, a histogram plot of unit spikes over time is created. In the second cell below, set stim_time to be the time of the stimulus you’re interested in viewing. To get an idea of the stimulus times you might be interested in, access one of the tables in the Intervals section, discussed above in Extracting Unit Data and Stimulus Data. The first cell below shows how to access these. Set interval_start and interval_end to the relative time bounds, in seconds, of the histogram around stim_time. Finally, start_unit and end_unit can be used to choose the slice indices of selected_units to display.
stim_time = 1007 # arbitrarily chosen here
interval_start = -0.5
interval_end = 5
start_unit = 100
end_unit = 500
spike_times = [elem for elem in units["spike_times"][start_unit:end_unit]]
if len(spike_times) == 0:
raise Exception("There are no spiking units in this selection")
len(spike_times)
400
# for each unit, generate a histogram with 275 bins, where bins represent the number spikes per second
time_bin_edges = np.linspace(interval_start, interval_end, 276)
hists = []
for unit_spike_times in spike_times:
hist, bins = np.histogram(unit_spike_times-stim_time, bins=time_bin_edges)
hists.append(hist)
hists = np.array(hists)
hists.shape
(400, 275)
# display array of histograms as 2D image with color
fig, ax = plt.subplots(figsize=(16,16))
img = ax.imshow(hists)
cbar = plt.colorbar(img, shrink=0.5)
cbar.set_label('# spikes')
ax.yaxis.set_major_locator(plt.NullLocator())
ax.set_ylabel("units", fontsize=16)
xtick_step=25
reltime = np.array(time_bin_edges)
ax.set_xticks(np.arange(0, len(reltime), xtick_step))
ax.set_xticklabels([f'{mp:1.1f}' for mp in reltime[::xtick_step]], rotation=45)
ax.set_xlabel("time since event (s)", fontsize=16)
ax.set_title("Units Spikes Over Time", fontsize=20)
Text(0.5, 1.0, 'Units Spikes Over Time')
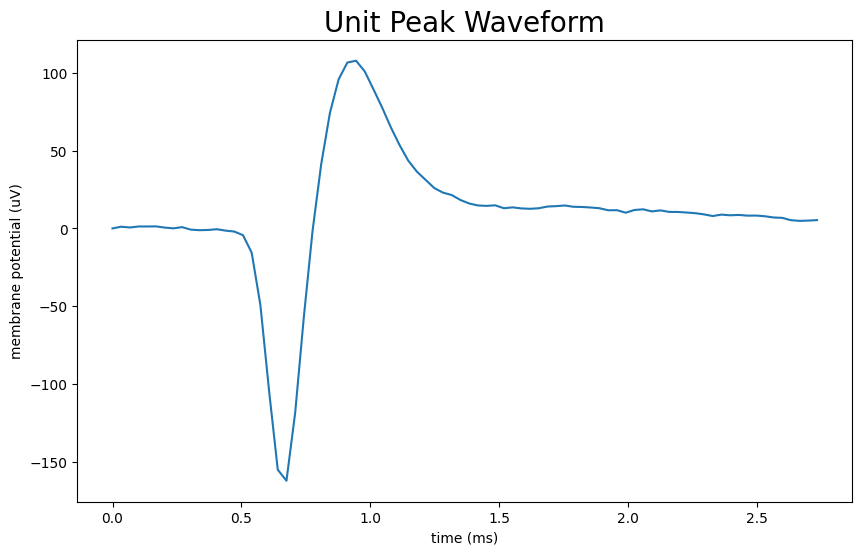
Visualizing Waveforms#
The Units table can also be used to view the waveforms of a units with the waveform_mean property, which consists of the mean waveform of that unit as measured by each channel along the probe. One channel will contain the peak waveform. With a bit of legwork, the peak_channel_id of the unit and the Electrodes table can be used to get the single peak waveform as shown below. Just as in the sections above, you don’t need to fully understand the Electrodes table except that it can be used to map channel IDs to probe IDs. There is also a timewise and channelwise view of all the mean waveforms and an average of the waveforms across all channels.
unit_num = 950952910
# translate unit id to row index
unit_idx = -1
for i in range(len(units.id)):
if units.id[i] == unit_num:
unit_idx = i
break
print("Unit index:",unit_idx)
Unit index: 2513
# get sampling Hz for this unit's waveform
peak_channel_id = units["peak_channel_id"][unit_idx]
electrodes = nwb.electrodes.to_dataframe()
probe_name = electrodes.loc[peak_channel_id].group_name
Hz = nwb.devices[probe_name].sampling_rate
Hz
29999.9995000281
Peak waveform#
# use peak_channel_id of unit to fetch the peak waveform index from electrodes table
peak_channel_id = units["peak_channel_id"][unit_idx]
electrodes = nwb.electrodes.to_dataframe()
local_index = electrodes.loc[peak_channel_id].local_index
unit_waveform = units["waveform_mean"][unit_idx]
peak_waveform = unit_waveform[local_index]
fig, ax = plt.subplots(figsize=(10,6))
n_secs = peak_waveform.shape[0] / Hz
time_axis = np.linspace(0, n_secs * 1000, peak_waveform.shape[0])
ax.plot(time_axis, peak_waveform)
ax.set_xlabel("time (ms)")
ax.set_ylabel("membrane potential (uV)")
ax.set_title("Unit Peak Waveform", fontsize=20)
plt.show()
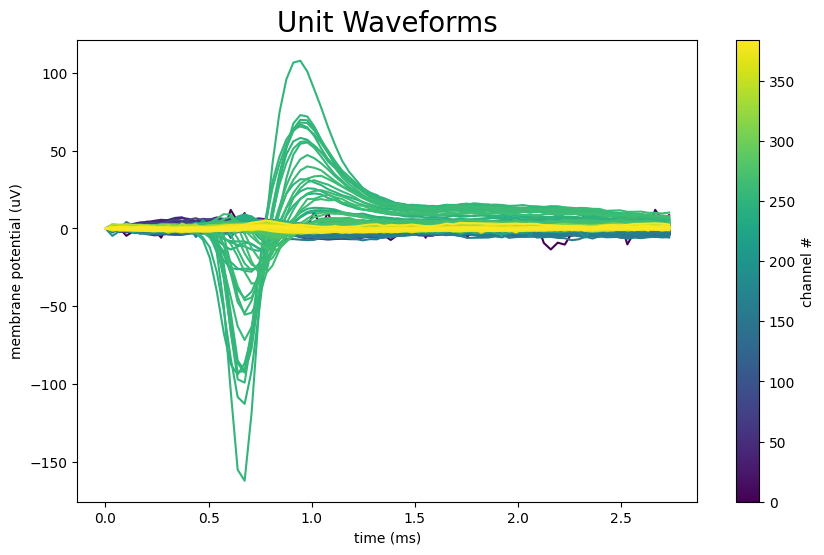
Waveforms#
Given the way Kilosort attributes spikes to specific units, most electrodes along the probe will detect nothing for a given unit. As the electrodes get closer to the actual source of the spikes, the waveform amplitude should increase and take shape. To get a very clear representation of a unit’s activity through space, the waveforms for a unit from all probes can be plotted. It can be seen that as the electrode gets further from the peak waveform, the amplitude decreases until the unit is too far away to be detected.
unit_waveforms = units["waveform_mean"][unit_idx]
unit_waveforms.shape
(384, 82)
fig, ax = plt.subplots(figsize=(10,6))
colors = plt.cm.viridis(np.linspace(0, 1, unit_waveforms.shape[0]))
ax.set_prop_cycle(color=colors)
n_secs = unit_waveforms.shape[1] / Hz
time_axis = np.linspace(0, n_secs * 1000, unit_waveforms.shape[1])
ax.plot(time_axis, np.transpose(unit_waveforms))
norm = mpl.colors.Normalize(vmin=0, vmax=len(colors))
cb = fig.colorbar(mpl.cm.ScalarMappable(norm=norm), ax=ax, label='channel #')
ax.set_xlabel("time (ms)")
ax.set_ylabel("membrane potential (uV)")
ax.set_title("Unit Waveforms", fontsize=20)
plt.show()
Waveform Image#
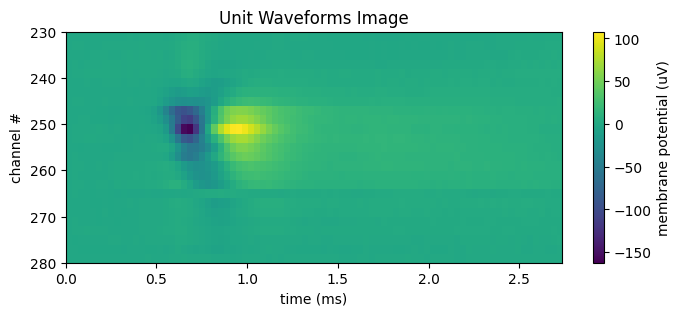
Below is an image of the waveform means for each channel of a unit. The further in space from the real neuron, the weaker the measurement of the response waveform, so it is usually only useful to view a subsection of the channels at once. Set start_channel and end_channel to be the bounds of the channels you want displayed. Because on neuropixels probes the channels are arranged into two rows along the length of a probe, typically a unit is only strongly detected by every other channel. The data shown below displays every other channel to avoid the resultant striping effect. If the waveform looks too dim, try incrementing start_channel by 1.
start_channel = 230
end_channel = 280
data = unit_waveforms[start_channel:end_channel:2] # step by 2 to remove striping effect
n_channels = end_channel - start_channel
fig, ax = plt.subplots(figsize=(8, n_channels // 15))
n_secs = unit_waveforms.shape[1] / Hz
time_axis = np.linspace(0, n_secs * 1000, unit_waveforms.shape[1])
norm = mpl.colors.Normalize(vmin=np.min(data), vmax=np.max(data))
cb = fig.colorbar(mpl.cm.ScalarMappable(norm=norm), ax=ax, label="membrane potential (uV)")
ax.set_xlabel("time (ms)")
ax.set_ylabel("channel #")
ax.set_title("Unit Waveforms Image")
ax.imshow(data, vmin=np.min(data), vmax=np.max(data), extent=[0, n_secs*1000, end_channel, start_channel], aspect="auto")
<matplotlib.image.AxesImage at 0x1a5326baf50>