Tutorial: Population Level Modeling (with PopNet)#
In this tutorial we will focus on modeling of populations and population firing rates. This is done with the PopNet simulator application of BMTK which uses the DiPDE engine as a backend. We will first build our networks using the BMTK NetworkBuilder and save them into the SONATA data format. Then we will show how to simulate the firing rates over a given interval.
Requirements:
BMTK
DiPDE
Scripts for running this tutorial can be found in the directory sources/chapter06.
For more information on BMTK and SONATA format, please see the Overview of BMTK and Network Builder pages.
1. Building the network#
Option 1: Converting existing networks#
Like BioNet for biophysically detailed modeling and PointNet with point-based networks, PopNet works with networks in the SONATA data format. PopNet supports simulating networks of individual cells at the population level. The first thing you have to do is modify the node-types and edge-types of an existing network to use Population level models (rather than models of individual cells).
WARNING - Converting a network of individual nodes into population of nodes is good for a quick and naive simulation, but for faster and more reliable results it’s best to build a network from scratch (next section).
See the SONATA users guide for details of a network set to work with BioNet (for an example of the BioNet network, see tutorial 3):
[1]:
import pandas as pd
pd.read_csv('sources/chapter06/converted_network/V1_node_types_bionet.csv', sep=' ')
[1]:
| node_type_id | ei | morphology_file | model_processing | pop_name | location | model_template | model_type | dynamics_params | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 100 | e | Scnn1a.swc | aibs_perisomatic | Scnn1a | L4 | ctdb:Biophys1.hoc | biophysical | 472363762_fit.json |
| 1 | 101 | i | Pvalb.swc | aibs_perisomatic | PV | L4 | ctdb:Biophys1.hoc | biophysical | 472912177_fit.json |
| 2 | 102 | e | NaN | NaN | LIF_exc | VisL4 | nrn:IntFire1 | point_process | IntFire1_exc_1.json |
| 3 | 103 | i | NaN | NaN | LIF_inh | VisL4 | nrn:IntFire1 | point_process | IntFire1_inh_1.json |
versus the equivalent form for PopNet:
[2]:
pd.read_csv('sources/chapter06/converted_network/V1_node_types_popnet.csv', sep=' ')
[2]:
| node_type_id | ei | morphology_file | model_processing | pop_name | location | model_template | model_type | dynamics_params | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 100 | e | NaN | NaN | Scnn1a | L4 | dipde:Internal | population | 472363762_pop.json |
| 1 | 101 | i | NaN | NaN | PV | L4 | dipde:Internal | population | 472912177_pop.json |
| 2 | 102 | e | NaN | NaN | LIF_exc | VisL4 | dipde:Internal | population | IntFire1_exc_pop.json |
| 3 | 103 | i | NaN | NaN | LIF_inh | VisL4 | dipde:Internal | population | IntFire1_inh_pop.json |
Some things to note:
Each node in the network now corresponds to a a population rather than a cell, and therefore model_type is node “population”, rather than individual biophysical/point types
We have set model_template to dipde:Internal which will tell the simulator to use special DiPDE model types
We are using new dynamic_params files with parameters that have been adjusted to appropiate range for DiPDE models.
morophology_file and model_processing, which were used to set and processes individual cell morphologies, are no longer applicable.
See SONATA specification for edges css files here. Finally, when we run the simulation we must tell PopNet to cluster nodes together using the group_by property:
network = popnet.PopNetwork.from_config(configure, group_by='node_type_id')
Option 2: Building a network from scratch#
We will create a network of two populations: one population of excitatory cells and another of inhibitory cells. Then we will save the network into SONATA-formatted data files.
The first step is to use the NetworkBuilder to instantiate a new network with two populations:
[3]:
from bmtk.builder import NetworkBuilder
net = NetworkBuilder('V1')
net.add_nodes(
pop_name='excitatory', # name of specific population optional
ei='e', # Optional
location='VisL4', # Optional
model_type='population', # Required, indicates what types of cells are being model
model_template='dipde:Internal', # Required, instructs what DiPDE objects will be created
dynamics_params='exc_model.json' # Required, contains parameters used by DiPDE during initialization of object
)
net.add_nodes(
pop_name='inhibitory',
ei='i',
model_type='population',
model_template='dipde:Internal',
dynamics_params='inh_model.json'
)
Next we will create connections between the two populations:
[4]:
net.add_edges(
source={'ei': 'e'}, target={'ei': 'i'},
syn_weight=0.005,
nsyns=20,
delay=0.002,
dynamics_params='ExcToInh.json'
)
net.add_edges(
source={'ei': 'i'}, target={'ei': 'e'},
syn_weight=-0.002,
nsyns=10,
delay=0.002,
dynamics_params='InhToExc.json'
)
[4]:
<bmtk.builder.connection_map.ConnectionMap at 0x7f5ea775c940>
and finally we must build and save the network:
[5]:
net.build()
net.save(output_dir='sim_ch06/network')
External Nodes#
The dipde:Internal nodes we created need input to fire, so we will use External Populations to drive the network activity. To do this we will create a separate network of ‘virtual’ populations, or alternativly use model_type=dipde:External, that connect to our excitatory population.
Note: we could add ‘virtual’ populations directly to our V1 network. However, creating them as a separate network provides a great advantage if/when we want to replace our external connections with a different model (or if we want to remove the reccurrent connections and simulate with feed-foward activity only).
[6]:
input_net = NetworkBuilder('LGN')
input_net.add_nodes(
pop_name='tON',
ei='e',
model_type='virtual'
)
input_net.add_edges(
target=net.nodes(ei='e'),
syn_weight=0.0025,
nsyns=10,
delay=0.002,
dynamics_params='input_ExcToExc.json'
)
input_net.build()
input_net.save(output_dir='sim_ch06/network')
2. Setting up the PopNet environment#
Before running the simulation we need to set up our simulation environment. This includes setting up run-scripts, configuration parameters, and placing our parameter files in their appropriate location. The easiest way to do this is through the command-line:
$ python -m bmtk.utils.sim_setup -n network --tstop 1500.0 popnet sim_ch06/
The function `bmtk.utils.sim_setup <https://alleninstitute.github.io/bmtk/_modules/bmtk/utils/sim_setup.html>`__ creates initial files to run a 1500 ms simulation (--tstop 1500.00) of the population network (popnet) using the network files found in our ./network directory (-n network) under the main directory (sim_ch06/).
Inputs#
We next need to set the firing rates of the External Population. There are multiple ways to set this value which will be discussed later. The best way is to set the firing rates using a input-rates file for each External Population. We can fetch an input-rates existing file using the command:
$ cd sim_ch06
$ wget https://github.com/AllenInstitute/bmtk/raw/develop/examples/pop_2pops/inputs/external_rates.csv -O lgn_rates.csv
Then we must open the simulation_config.json file with a text editor and add the lgn_rates.csv file as a part of our inputs:
{
"inputs": {
"LGN_pop_rates": {
"input_type": "csv",
"module": "pop_rates",
"rates": "${BASE_DIR}/lgn_rates.csv",
"node_set": "LGN"
}
}
}
3. Running the simulation#
The call to sim_setup created a file run_pointnet.py that we can run directly in a command line:
$ python run_popnet.py config.json
It takes one argument, config.json, which contains the simulation parameters specified during the previous steps.
Alternatively, we can run it directly using the following python code:
[7]:
from bmtk.simulator import popnet
configure = popnet.config.from_json('sim_ch06/config.json')
configure.build_env()
network = popnet.PopNetwork.from_config(configure)
sim = popnet.PopSimulator.from_config(configure, network)
sim.run()
2022-08-09 16:50:15,020 [INFO] Created log file
2022-08-09 16:50:15,054 [INFO] Building cells.
2022-08-09 16:50:15,055 [INFO] Building recurrent connections
2022-08-09 16:50:15,059 [INFO] Build virtual cell stimulations for LGN_pop_rates
2022-08-09 16:50:15,078 [INFO] Network created.
2022-08-09 16:50:15,080 [INFO] Running simulation.
2022-08-09 16:50:18,766 [INFO] Finished simulation.
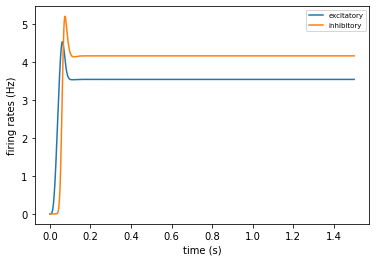
4. Analyzing the results#
As specified in the “output” section of the simulation_config.json file, the results will be written to ouput/spike_rates.csv. The BMTK analyzer includes code for plotting and analyzing the firing rates of our network:
[8]:
from bmtk.analyzer.firing_rates import plot_rates_popnet
plot_rates_popnet('sim_ch06/network/V1_node_types.csv', 'sim_ch06/output/firing_rates.csv', model_keys='pop_name')
[ ]: