BMTK: The Brain Modeling Toolkit#
About BMTK#
The Brain Modeling Toolkit (BMTK) is an open-source software package for modeling and simulating large-scale neural network models. It supports a range of modeling resolutions, including multi-compartment, biophysically detailed models, point-neuron models, and population-level firing rate models.
BMTK provides a full workflow for developing biologically realistic brain network models—from building networks from scratch, to running parallelized simulations, to conducting perturbation analyses. Its features include:
A unified interface (API) for building and running simulations across levels of resolution
A flexible framework for sharing models and expanding upon existing ones
A simple simulation setup requiring little-to-no programming
Adaptability that allows advanced users to fully customize how networks are instantiated and simulated
Automatic parallelization
Support for simulations ranging from single cells to networks with millions of cells and billions of synapses
On-the-fly adjustments of cell and synaptic properties during simulations
A suite of functions for analyzing and visualizing network structure and simulation results
And much more!

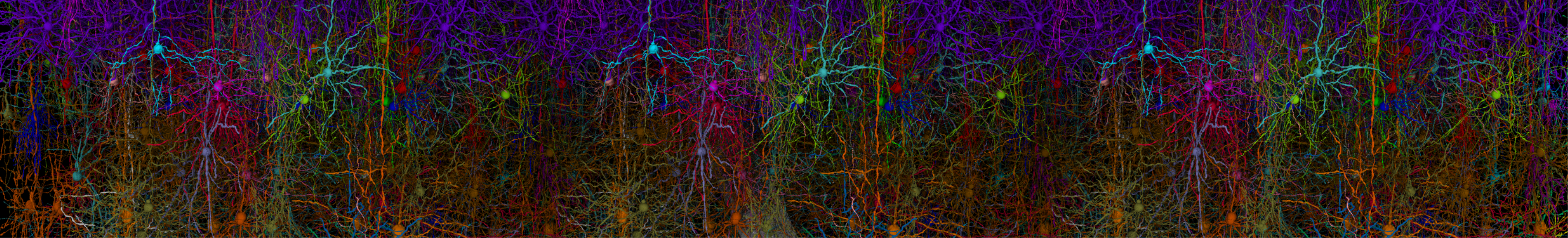
Simulation of the mouse primary visual cortex, prepared and carried out using BMTK.#
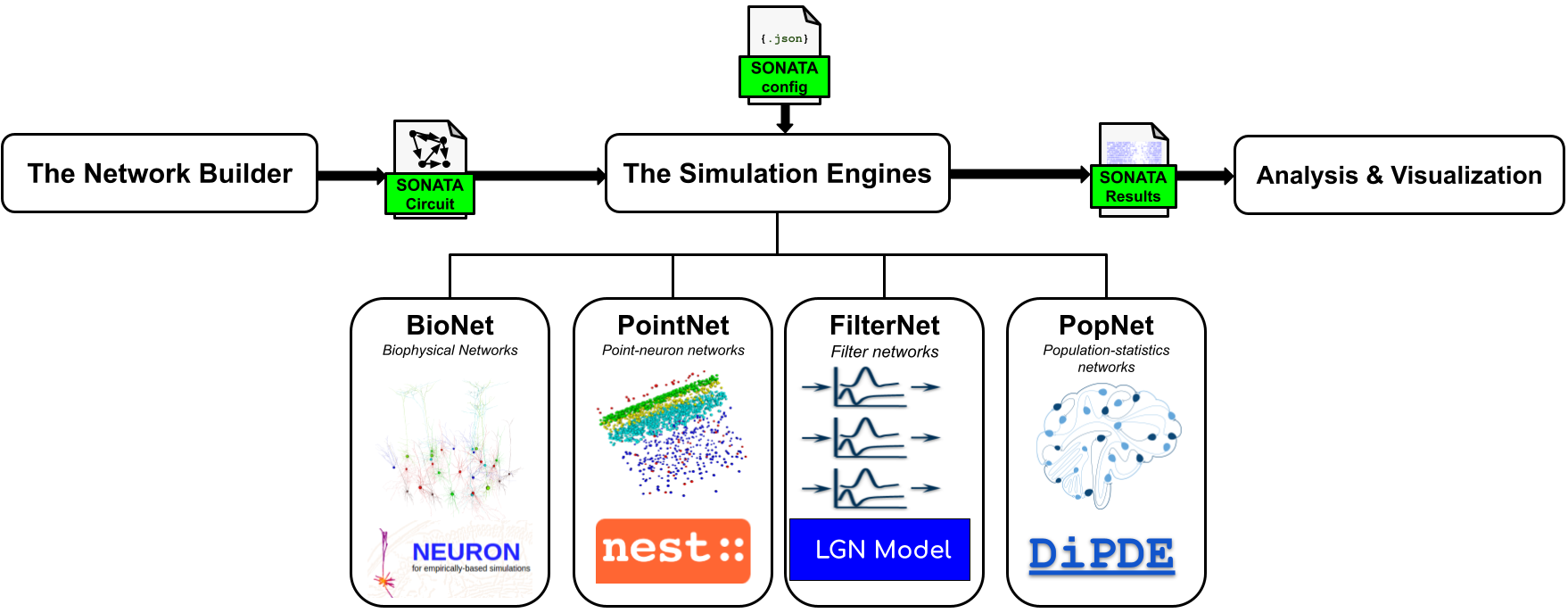
BMTK Workflow
BMTK uses a modular organization and the SONATA data format (SONATA, see below) to separate the processes of building, simulating, and analyzing brain network models. BMTK constructs a fully instantiated model and saves it in SONATA files. Then, running a simulation simply involves loading those SONATA files, with no need to rebuild the model each time.
Users can also adjust cell and synaptic parameters in the SONATA files to iterate more quickly on simulations. Models in SONATA format can be constructed, simulated, or analyzed not only with BMTK but also with other tools that support the format.
BMTK consists of three major components: the network builder, the simulation engines, and the analysis and visualization tools. The components can be used in one workflow or separately.
Further Resources#
For detailed information about using BMTK and all the available features please see our User Guide page.
For a list of workable tutorials and example networks please see our Tutorials and Examples page.
For examples of how we use BMTK and related tools to build realistic models at the Allen Institute, please see our Computational Modeling & Theory page at the Allen Brain Map Portal.
If you have questions, issues, or requests for BMTK developers and/or the scientists that use the tool in their own modeling work, please feel free to reach out to us directly. Our Contact Us page contains a number of ways to reach out to our team at the Allen Institute.
Acknowledgements#
How to cite BMTK and related tools.
See our Contributors Page for a list of the people who have helped with the development and growth of BMTK.
We wish to thank the Allen Institute founder, Paul G. Allen, for his vision, encouragement, and support.