Synaptic plasticity in Pointnet (STP, facilitation, depression, STDP, and others)¶
In the NEST simulator, the available postsynaptic current or conductance waveforms (alpha, exp, or delta) are typically integrated into the neuron model. The synapse model allows for more sophisticated and varied built-in models of synapses, such as dynamic synapses or gap junctions.
A list of available synapse models is listed here: https://nest-simulator.readthedocs.io/en/v2.18.0/models/synapses.html
So far, we have been using “static_synapse” (which simply stores target, weight, delay and receiver port). In this mini-tutorial, we will show an example of how to implement facilitation and depression using BMTK. This approach can be used to access other synaptic models as well.
[1]:
import matplotlib.pyplot as plt
import nest
import numpy as np
import os
import glob
from bmtk.builder.networks import NetworkBuilder
from bmtk.simulator import pointnet
from bmtk.analyzer.compartment import plot_traces
from bmtk.analyzer.spike_trains import plot_raster
-- N E S T --
Copyright (C) 2004 The NEST Initiative
Version: nest-3.0
Built: Sep 14 2022 22:43:51
This program is provided AS IS and comes with
NO WARRANTY. See the file LICENSE for details.
Problems or suggestions?
Visit https://www.nest-simulator.org
Type 'nest.help()' to find out more about NEST.
First, we will create a two neuron network. The presynaptic neuron is a simple integrate-and-fire neuron. If we open the dynamics_params file 472363762_point_with_cc.json within /components/point_neuron_models, we see that the parameter I_e has been set to 190.0, which injects a constant background current, triggering regular firing in the presynaptic neuron.
{
"I_e": 190.0,
"tau_m": 44.9,
"C_m": 239.0,
"t_ref": 3.0,
"E_L": -78.0,
"V_th": -43.0,
"V_reset": -55.0
}
[2]:
net = NetworkBuilder('two_cell')
# Build Nodes
net.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:iaf_psc_alpha',
dynamics_params='472363762_point_with_cc.json',
ei_type='e',
pop_name='presyn'
)
net.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:glif_lif_asc_psc',
dynamics_params='Scnn1a_515806250_glif_lif_asc.json',
ei_type='e',
pop_name='postsyn'
)
We will add a ‘static_synapse’ connection between the neurons so we can first see the baseline behavior of the network with an inhibitory static synapse.
[3]:
# Build edge
net.add_edges(
source=net.nodes(pop_name='presyn'),
target=net.nodes(pop_name='postsyn'),
connection_rule = 1,
model_template='static_synapse',
dynamics_params='static_ExcToExc.json',
delay=2.0,
syn_weight=-100
)
net.build()
net.save(output_dir='sim_dyn_syn/network')
[4]:
from bmtk.utils.create_environment import create_environment
create_environment(
'pointnet',
base_dir='sim_dyn_syn',
config_file='config.pointnet.json',
components_dir='../sources/pointnet_files/dynamic_synapses',
network_dir='network',
output_dir='output_pointnet',
tstop=3000.0, dt=0.1,
report_vars=['V_m'],
run_script='run_pointnet.py',
overwrite=True,
)
[5]:
configure = pointnet.Config.from_json('sim_dyn_syn/config.pointnet.json')
configure.build_env()
network = pointnet.PointNetwork.from_config(configure)
sim = pointnet.PointSimulator.from_config(configure, network)
sim.run()
2022-10-28 17:26:43,972 [INFO] Created log file
INFO:NestIOUtils:Created log file
2022-10-28 17:26:43,986 [INFO] Batch processing nodes for two_cell/0.
INFO:NestIOUtils:Batch processing nodes for two_cell/0.
2022-10-28 17:26:43,998 [INFO] Setting up output directory
INFO:NestIOUtils:Setting up output directory
2022-10-28 17:26:44,000 [INFO] Building cells.
INFO:NestIOUtils:Building cells.
2022-10-28 17:26:44,006 [INFO] Building recurrent connections
INFO:NestIOUtils:Building recurrent connections
2022-10-28 17:26:44,012 [INFO] Network created.
INFO:NestIOUtils:Network created.
2022-10-28 17:26:44,018 [INFO] Starting Simulation
INFO:NestIOUtils:Starting Simulation
2022-10-28 17:26:44,159 [INFO] Simulation finished, finalizing results.
INFO:NestIOUtils:Simulation finished, finalizing results.
2022-10-28 17:26:44,217 [INFO] Done.
INFO:NestIOUtils:Done.
[6]:
# Presynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [0])
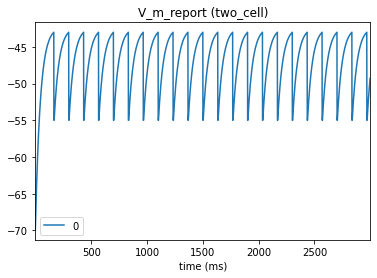
Our background current produces regular firing in the presynaptic cell. Note that for LIF models the spike itself is not shown and just produces a reset of the membrane potential.
[7]:
# Postsynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [1])
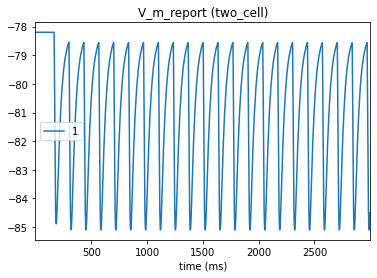
In the postsynaptic cell, we can see the summation of a series of IPSPs triggered by each presynaptic spike (note that the postsynaptic effect is modeled as a current but produces the voltage effect seen here).
Next, we’ll try rebuilding the network, this time with a facilitating synapse based on the “tsodyks2_synapse” model. This model is described in more detail at:
https://nest-simulator.readthedocs.io/en/stable/auto_examples/evaluate_tsodyks2_synapse.html
as well as on the synapses documentation page linked at the top.
[9]:
# Clear the network folder
for f in glob.glob('sim_dyn_syn/network/*'):
try:
os.remove(f)
except FileNotFoundError as fnfe:
pass
[10]:
net2 = NetworkBuilder('two_cell_fac')
# Build Nodes
net2.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:iaf_psc_alpha',
dynamics_params='472363762_point_with_cc.json',
ei_type='e',
pop_name='presyn'
)
net2.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:glif_lif_asc_psc',
dynamics_params='Scnn1a_515806250_glif_lif_asc.json',
ei_type='e',
pop_name='postsyn'
)
[11]:
net2.add_edges(
source=net2.nodes(pop_name='presyn'),
target=net2.nodes(pop_name='postsyn'),
connection_rule = 1,
model_template='tsodyks2_synapse',
dynamics_params='TsodyksFac_glif.json',
delay=2.0,
syn_weight=-100
)
net2.build()
net2.save(output_dir='sim_dyn_syn/network')
We have created a new dynamics_params file tsodyksFac_glif.json based on the dynamic_params file for the static synapse and have added to it the following params for the tsodyks2 synapse:
{
"U": 0.1,
"u": 0.1,
"x": 1.0,
"tau_rec": 100.0,
"tau_fac": 1000.0,
"weight": 250.0,
}
[12]:
create_environment(
'pointnet',
base_dir='sim_dyn_syn',
config_file='config.pointnet.json',
components_dir='../sources/pointnet_files/dynamic_synapses',
network_dir='network',
output_dir='output_pointnet',
tstop=3000.0, dt=0.1,
report_vars=['V_m'],
run_script='run_pointnet.py',
overwrite=True,
)
[13]:
configure = pointnet.Config.from_json('sim_dyn_syn/config.pointnet.json')
configure.build_env()
network = pointnet.PointNetwork.from_config(configure)
sim = pointnet.PointSimulator.from_config(configure, network)
sim.run()
2022-10-28 17:27:28,710 [INFO] Created log file
INFO:NestIOUtils:Created log file
2022-10-28 17:27:28,728 [INFO] Batch processing nodes for two_cell_fac/0.
INFO:NestIOUtils:Batch processing nodes for two_cell_fac/0.
2022-10-28 17:27:28,739 [INFO] Setting up output directory
INFO:NestIOUtils:Setting up output directory
2022-10-28 17:27:28,740 [INFO] Building cells.
INFO:NestIOUtils:Building cells.
2022-10-28 17:27:28,746 [INFO] Building recurrent connections
INFO:NestIOUtils:Building recurrent connections
2022-10-28 17:27:28,756 [INFO] Network created.
INFO:NestIOUtils:Network created.
2022-10-28 17:27:28,763 [INFO] Starting Simulation
INFO:NestIOUtils:Starting Simulation
2022-10-28 17:27:28,878 [INFO] Simulation finished, finalizing results.
INFO:NestIOUtils:Simulation finished, finalizing results.
2022-10-28 17:27:28,924 [INFO] Done.
INFO:NestIOUtils:Done.
[14]:
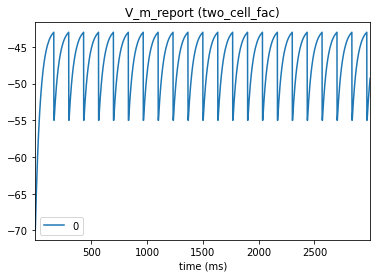
# Presynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [0])
[15]:
# Postsynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [1])
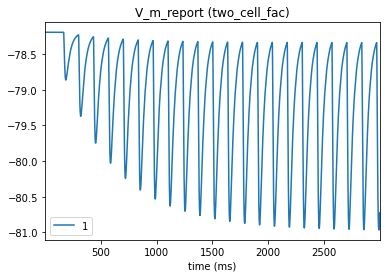
This time, the IPSP’s get incrementally larger as they occur closely in succession, simulating synaptic facilitation. We can use the same model, just changing the parameters, to create a depressing synapse. We will add these parameter values to the dynamic_params:
{
"U": 0.67,
"u": 0.67,
"x": 1.0,
"tau_rec": 450.0,
"tau_fac": 0.0,
"weight": 250.0
}
[16]:
# Clear the network folder
for f in glob.glob('sim_dyn_syn/network/*'):
try:
os.remove(f)
except FileNotFoundError as fnfe:
pass
[17]:
net3 = NetworkBuilder('two_cell_dep')
# Build Nodes
net3.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:iaf_psc_alpha',
dynamics_params='472363762_point_with_cc.json',
ei_type='e',
pop_name='presyn'
)
net3.add_nodes(
N=1,
model_type='point_neuron',
model_template= 'nest:glif_lif_asc_psc',
dynamics_params='Scnn1a_515806250_glif_lif_asc.json',
ei_type='e',
pop_name='postsyn'
)
[18]:
net3.add_edges(
source=net3.nodes(pop_name='presyn'),
target=net3.nodes(pop_name='postsyn'),
connection_rule = 1,
model_template='tsodyks2_synapse',
dynamics_params='TsodyksDep_glif.json',
delay=2.0,
syn_weight=-100
)
net3.build()
net3.save(output_dir='network')
[19]:
create_environment(
'pointnet',
base_dir='sim_dyn_syn',
config_file='config.pointnet.json',
components_dir='../sources/pointnet_files/dynamic_synapses',
network_dir='network',
output_dir='output_pointnet',
tstop=3000.0, dt=0.1,
report_vars=['V_m'],
run_script='run_pointnet.py',
overwrite=True,
)
[20]:
configure = pointnet.Config.from_json('sim_dyn_syn/config.pointnet.json')
configure.build_env()
network = pointnet.PointNetwork.from_config(configure)
sim = pointnet.PointSimulator.from_config(configure, network)
sim.run()
2022-10-28 17:27:54,260 [INFO] Created log file
INFO:NestIOUtils:Created log file
2022-10-28 17:27:54,272 [INFO] Batch processing nodes for two_cell_dep/0.
INFO:NestIOUtils:Batch processing nodes for two_cell_dep/0.
2022-10-28 17:27:54,282 [INFO] Setting up output directory
INFO:NestIOUtils:Setting up output directory
2022-10-28 17:27:54,283 [INFO] Building cells.
INFO:NestIOUtils:Building cells.
2022-10-28 17:27:54,289 [INFO] Building recurrent connections
INFO:NestIOUtils:Building recurrent connections
2022-10-28 17:27:54,294 [INFO] Network created.
INFO:NestIOUtils:Network created.
2022-10-28 17:27:54,299 [INFO] Starting Simulation
INFO:NestIOUtils:Starting Simulation
2022-10-28 17:27:54,437 [INFO] Simulation finished, finalizing results.
INFO:NestIOUtils:Simulation finished, finalizing results.
2022-10-28 17:27:54,485 [INFO] Done.
INFO:NestIOUtils:Done.
[21]:
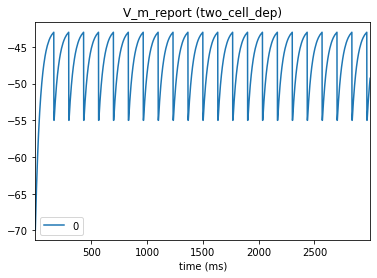
# Presynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [0])
[22]:
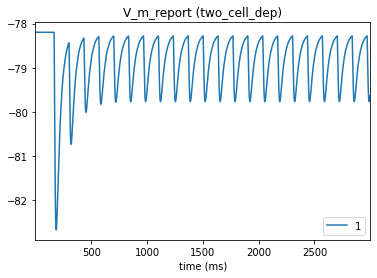
# Postsynaptic cell
_ = plot_traces(config_file='sim_dyn_syn/config.pointnet.json', report_name='V_m_report', node_ids = [1])
[ ]:
