Getting started#
Let us start by selecting the human atlas as a test case. All data are stored on a public S3 bucket:
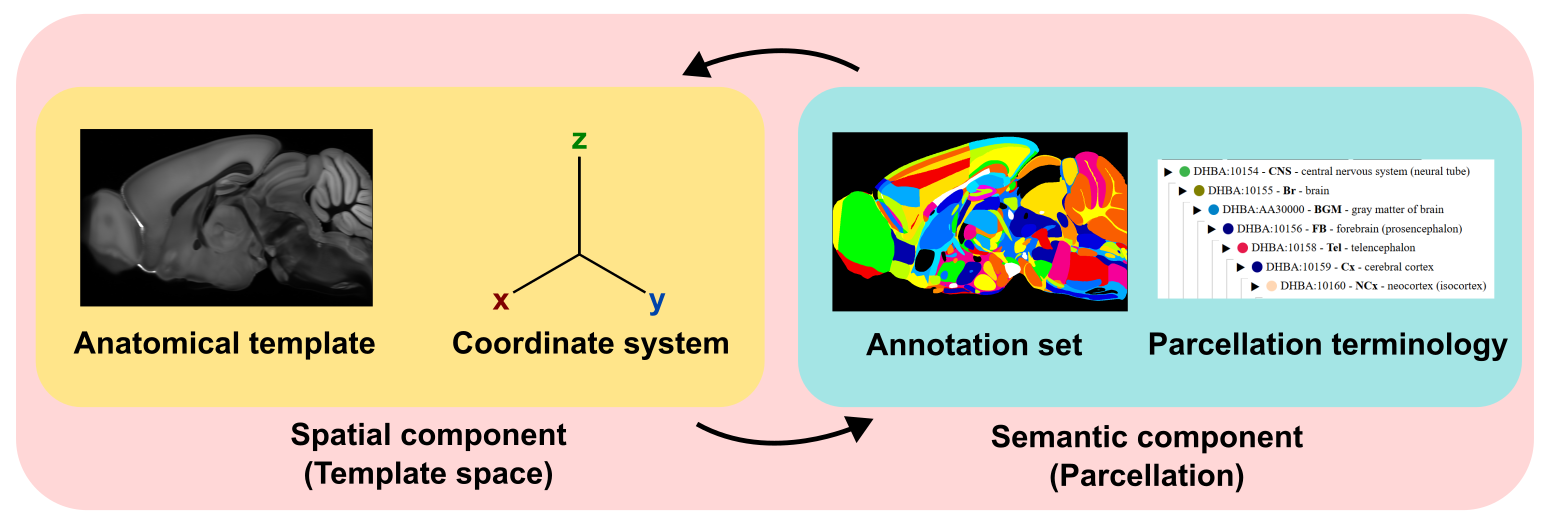
All atlases have three major components: the template, annotation, and terminology
The template is a reference image volume that defines an coordinate space. The coordinate space sets the orientation and direction of the x,y,z axes and defines the voxel size. In the macaque case, the anatomical template is the Mac25Rhesus template, which is comprised of an averaged T1 weighted MRI from 25 adult rhesus macaques and resampled to 160 um voxel resolution.
The annotation is a set of manually defined segmentations of the template space. These segmentations parcellate the template into defined structures. Each segmentation is assigned a unique numeric value.
Annotations are named and labeled using the Harmonized Ontology of Mammalian Brain Anatomy (HOMBA), which provides a terminology, a taxonomy of the structures and their relationship along with relevant metadata (eg. label colors), and serves as a lookup between the annotation value and the label.
Together, these components uniquely define an anatomical atlas
For a deeper understanding of atlases and their components, see the atlas documentation
Install notebooks and dependencies#
If you haven’t already installed necessary dependencies from the github repository, we will do so here using a terminal. These dependencies are prerequisites for running notebooks. It is strongly advised to install these into a clean conda environment or other virtual environment (eg. uv).
This installs all necessary dependencies. However, if we want to use the notebooks locally (including this one), we will need to get them from the github repository as a zip file.
To run this notebook, navigate to the unzipped directory. In the terminal, type
jupyter notebook
This should open your browser. Navigate to the /notebooks directory to view and run the notebooks
Download all data assets associated with an atlas#
Individual files can be downloaded manually (see Data section). However, we recommend using the atlas manifest to download the combined template, annotation, and terminology components as a whole. The manifest contains relative S3 paths to all data assets and is used to download data to a local directory. The current manifest version for HMBA atlases is 2025.
import atlas_utils.atlas_import as atlas
atlas.get_latest_atlases()
Downloaded data\atlases\allen-adult-mouse-ccf-stereotaxic-atlas\2020\manifest.json
Downloaded data\atlases\hmba-adult-human-homba-atlas\2025\manifest.json
Downloaded data\atlases\hmba-adult-human-hombabg-atlas\2025\manifest.json
Downloaded data\atlases\hmba-adult-macaque-homba-atlas\2025\manifest.json
Downloaded data\atlases\hmba-adult-marmoset-homba-atlas\2025\manifest.json
We can now use the local manifest files to download packaged atlases. We currently support the HMBA atlases for human, macaque, and marmoset. Selecting all downloads all atlases to the local machine.
Note that there are currently two human atlases, a whole brain gyral and modified Brodmann parcellation in MNI ICBM 2009b space (human-homba) and a basal ganglia parcellation in HCP space (human-hombabg). These can be accessed by downloading either human-mni or human-hcp
atlas.download_atlas('human-hcp')
Destination directory: C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025
C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025\annotations_compressed_700.nii.gz
C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025\parcellation_volumes.csv
C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025\template_700.nii.gz
C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025\HOMBA_v1_mapping.csv
C:\Users\ashwin.bhandiwad\Documents\CCF_MAP\notebooks\data\atlases\hmba-adult-human-hcp-homba-atlas\2025\terminology.csv
Next steps#
For notes on using the annotation for labeling and indexing data, see Working with annotation volumes
