Consensus Basal Ganglia Cell Type Atlas: Annotation Coordination Hub
This repository is a central resource for annotators collaborating on a cross-species consensus atlas of basal ganglia cell types. Our collaborative goal is to integrate molecular, anatomical, and developmental insights to refine basal ganglia cell type taxonomies in human, macaque, marmoset and mouse.
Goals of the Annotation Effort:
- Establish a consensus cell type nomenclature across species.
- Identify robust marker genes (including transcription factors and neurotransmitters) and assess conservation across species.
- Clarify neuronal diversity through anatomical and developmental characterization.
- Reorganize and refine clusters into biologically meaningful hierarchical groups.
How to Participate:
- Create a profile for the Cell Annotation Platform (CAP).
- Load the CAP atlas and click on “Molecular Data”.
- Provide feedback on annotations in the Molecular Data Overview.
Additional Resources:
- CAP Orientation Videos led by Evan Biederstedt.
- Cell type metadata including marker genes and anatomical distributions.
Cell Annotation Platform (CAP)
To facilitate collaborative annotation with members of the community we are using the Cell Annotation Platform (CAP) to put our work into the hands of researchers who can provide feedback, thoughts and expert information about the consensus nomeclature system being proposed.
We have provided the Consensus Cross-species Taxonomy as an integrated object containing nuclei/cells from Human, Macaque and Marmoset as a complete Atlas and various sub-groupings of taxonomic terms to provide a higher-resolution view for each Class of cell types.
If you have any questions or requests please reach out to either Nelson Johansen (nelson.johansen@alleninstitute.org) or Trygve Bakken (trygveb@alleninstitute.org).
Conserved Marker Analysis
Computational identificaiton of conserved marker genes across species will be provided shortly.
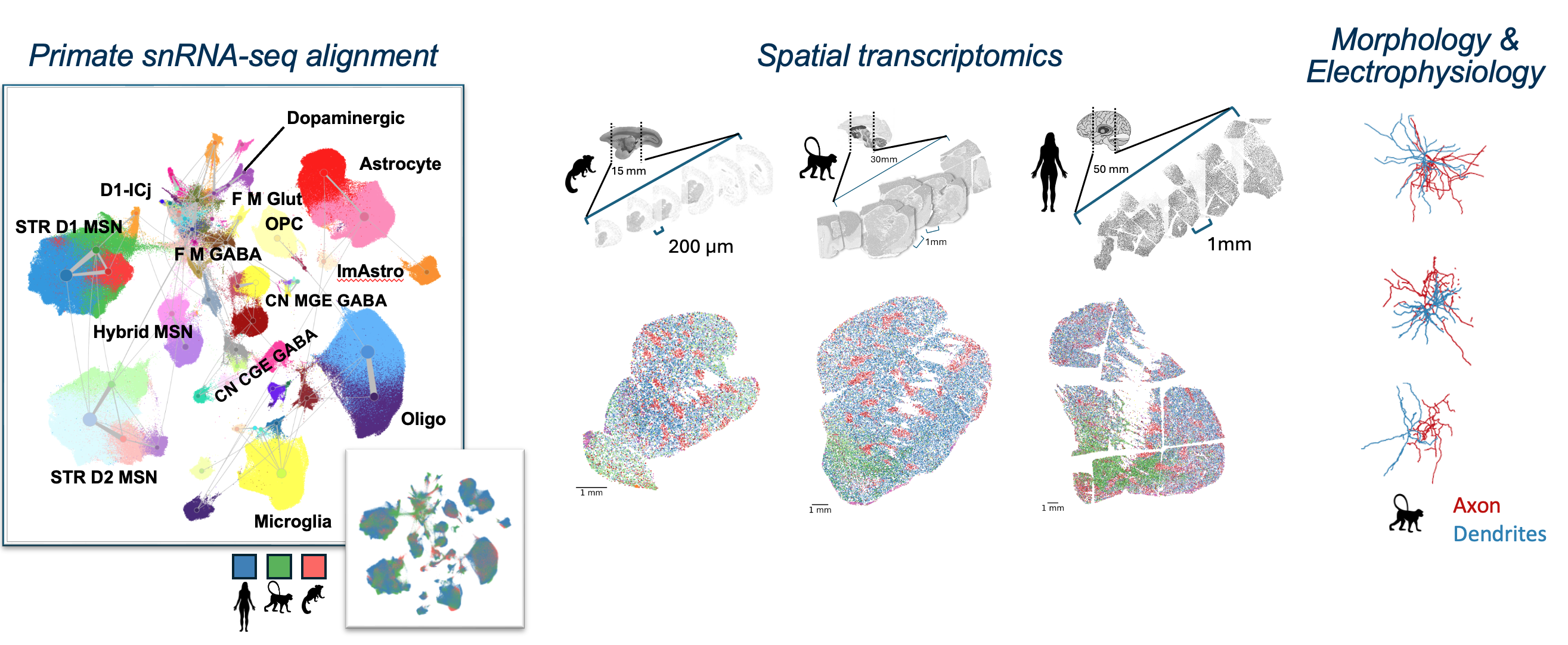
Spatial transcriptomic cirrocumulus
We have provided a cirrocumulus browser for access to the Macaque spatial transcriptomics data available here https://cirro-aibs.org/.
- Login with user: hmbajamboree@gmail.com; password: welcomeannotators
- Click Open (near top right) and chose any of:
- Macaque BG Neurons for Jamboree – final taxonomy (~1M cells)
- Macaque BG Non-neurons for Jamboree – final taxonomy (~6.2M cells)
- Marmoset BG Jamboree (and then open the
spatial_cirro_gridembedding in the top left – 2.2M cells)
Performance may be slow, please have patience.
Compare cell type annotations and metadata
Annotation Comparison Explorer (ACE) provides interactive visualizations comparing basal ganglia cell type assignments with alignments to the adult human brain atlas (Siletti et al 2023), the adult mouse brain atlas (Yao et al 2023), and other metadata such as anatomic structure, species, and donor sex.
- Visit this site for details, including ACE orientation videos for this data set
- Access ACE directly here: https://sea-ad.shinyapps.io/ACEapp/
Coordination Team:
Trygve Bakken, Nelson Johansen, Yuanyuan Fu, Tyler Mollenkopf, Jeremy Miller, Lauren Alfiler, Yasmeen Hussain and Julie Nyhus (Allen Institute for Brain Science)
Supported by NIH Cell Atlas Network grant UM1MH130981, the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NIH) under award number U24NS133077, and the NIH BRAIN Initiative under award number U24MH130918.




